4237
Standard DICOM Structured Reports as a Vehicle for Multi-Modal Region-of-Interest MR Analysis Results for Clinical Workflows and Radiomic Studies for Brain Tumor Patients1Radiology and Biomedical Imaging, UCSF, San Francisco, CA, United States
Synopsis
Quantitative analysis of metabolic and dynamic imaging data produces maps of parameters that show promise for improving medical diagnosis and therapeutic monitoring for patients with brain tumors. Statistical ROI analysis of these maps can be used to quantitatively summarize multi-modality imaging metrics and longitudinal changes. In this work we demonstrate a standards-based mechanism for generating and communicating minable, quantitative Region of Interest (ROI) analysis results that can easily be integrated into clinical workflows and radiomic studies.
Purpose
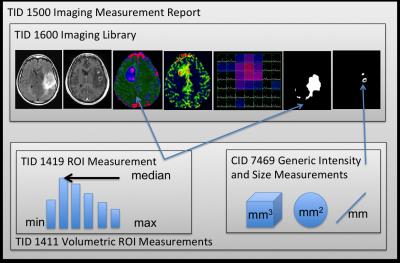
To demonstrate a standards-based mechanism for generating and communicating minable, quantitative Region-of-Interest (ROI) analysis results that can easily be integrated into clinical workflows and radiomic studies. Quantitative analysis of metabolic and dynamic imaging data produces maps of parameters that show promise for improving medical diagnosis and therapeutic monitoring for patients with brain tumors1. Statistical ROI analysis of these maps can be used to quantitatively summarize multi-modality imaging metrics and longitudinal changes. Such metrics are important for radiomic studies, which aim to characterize radiological data based on sets of image features2. DICOM Secondary Capture reports can be used to send visual reports containing such quantitative results to PACS, but it is desirable to a) capture quantitative results in a machine-readable format that can be stored with the clinical record and b) annotate the results using standardized terminology. The DICOM Structured Report (SR)3 is designed to store and communicate structured data, but requires a template to define domain specific content. Here we report tools implemented within the SIVIC4 software package to read, write and visualize DICOM SR to facilitate incorporation of quantitative MR metrics in clinical workflows and radiomic studies. A brain-tumor SR template (btSR) was derived here from a template developed for PET/CT head and neck cancer5. New terms for annotating brain tumor ROI data were identified where terms in existing dictionaries (SNOMED, SRT, DCM, etc)6 did not exist.Methods
SIVIC is an open-source software framework and application suite for the evaluation and interpretation of MRSI and dynamic imaging data. It leverages a number of free open-source packages including VTK7, ITK8 and DCMTK9. We have recently expanded SIVIC to perform statistical ROI analysis. This functionality is implemented in the svkImageStatistics class, which is a compilation of existing statistics methods from the ITK and VTK toolkits as well as new methods such as the computation of quantiles from smoothed histograms. The DCMTK DCMSR module was used to export this data into DICOM SR conforming to the btSR schema. Multi-modal MR exams for brain tumor patients were used to test our workflow. 3D DICOM parameter maps representing the apparent diffusion coefficient, cerebral blood volume, and choline to NAA index (CNI)10 were derived using open-source and in-house processing software. ROIS representing the contrasting enhancing lesion, T2 hyperintense lesion, necrotic region, and surgical cavity were manually drawn and exported as DICOM Segmentation objects11. An ROI with CNI > 2 (CNI2) was defined automatically from the spectral data. The svk_image_stats command line tool was used to compute many statistical measures including the median, maximum, standard deviation, 25th, and 75th percentiles for pixel intensities of images and parameter maps within each ROI. Geometric measurements were generated for each ROI including volume, surface area, greatest within-slice cross-section and greatest overall cross-section in 3D.Results
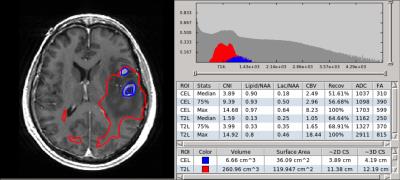
All of the results from the analysis were stored as a DICOM SR that conformed to our btSR (figure 1). Researchers were able to view the data through the SIVIC GUI and gave feedback regarding the presentation of the results (figure 2). For serial data analysis the GUI supports the comparison of metrics at multiple timeponts. Results were stored in our research PACS along with the complete DICOM record.Discussion
Incorporation of quantitative MRI results into clinical workflows and radiomic studies has been limited by the absence of adopted standards. The DICOM btSR developed here provides a standard that could be used to overcome this limitation. One of the practical issues that remains is that although many PACS can store SR data they cannot visualize it. The SIVIC GUI provides an interactive interface for visualizing this information and is also capable of interactively generating DICOM SC reports that can also be sent to PACS to display summary quantitative results. Radiomic studies in contrast only require a well-documented machine-readable format, which the btSR provides.Conclusion
The DICOM btSR template developed here allowed us to store, communicate, and analyze quantitative multi-modal ROI analysis results for serial data from patients with brain tumors using standard DICOM infrastructure. The SR format provides a mechanism for including quantitative imaging results in clinical workflows and for sharing data in multi-center trials. A number of terms required to annotate the brain-tumor ROI metrics did not exist in standard dictionaries and will be proposed for addition to the DICOM standard. Using the SIVIC GUI to load the quantitative results along with the relevant images and ROIs significantly improves the time to review and analyze the data thus reducing the additional overhead require to integrate this type of data into research and clinical workflows.Acknowledgements
This work was supported by: P01 CA118816References
1. Saraswathy, S. et al. Evaluation of MR markers that predict survival in patients with newly diagnosed GBM prior to adjuvant therapy. J. Neurooncol. 91, 69–81 (2009).
2. Lambin, P. et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 48, 441–446 (2012).
3. Clunie, D. DICOM structured reporting. (2000). Available at: http://www.dclunie.com/pixelmed/DICOMSR.book.pdf.
4. SIVIC. Available at: http://sivic.sourceforge.net.
5. Fedorov, A. et al. DICOM for quantitative imaging biomarker development: a standards based approach to sharing clinical data and structured PET/CT analysis results in head and neck cancer research. PeerJ 4, e2057 (2016).
6. Bidgood, W. D. The SNOMED DICOM microglossary: Controlled terminology resource for data interchange in biomedical imaging. Methods Inf. Med. 37, 404–414 (1998).
7. VTK. Available at: http://www.vtk.org.
8. ITK. Available at: https://itk.org.
9. DCMTK. Available at: http://dicom.offis.de/dcmtk.php.en.
10. McKnight, T. R., Noworolski, S. M., Vigneron, D. B. & Nelson, S. J. An automated technique for the Quantitative assessment of 3D-MRSI data from patients with glioma. J. Magn. Reson. Imaging 13, 167–177 (2001).
11. DICOM Segmentation. Available at: ftp://medical.nema.org/medical/dicom/final/sup111_ft.pdf.
Figures