4124
Towards automated MP2RAGE-based hippocampal volumetry for routine clinical practiceBénédicte Maréchal1,2,3, Sophie Espinoza4, Wadie Ben Hassen5, Tobias Kober1,2,3, Christophe Habas4, and Alexis Roche1,2,3
1Advanced Clinical Imaging Technology, Siemens Healthcare HC CEMEA SUI DI PI, Lausanne, Switzerland, 2Department of Radiology, CHUV, Lausanne, Switzerland, 3LTS5, EPFL, Lausanne, Switzerland, 4Service de NeuroImagerie, Centre Hospitalier National d'Ophtalmologie des XV-XX, Paris, France, 5Siemens Healthcare S.A.S., Saint-Denis, France
Synopsis
We investigate the potential of an automated method to provide hippocampal volume estimates using T1-weighted images obtained from the MP2RAGE sequence. Scheltens visual medial temporal atrophy scores from 27 patients undergoing clinical brain MRI for workup of cognitive decline were compared to automated volumetric measures using multinomial logistic regression. Strong correlation was observed which suggests that the employed hippocampal volumetry method may help supporting diagnosis of neurodegenerative diseases.
Purpose
Hippocampal volumetry is becoming one of the most established and validated diagnostic markers for Alzheimer’s disease [1,2]. Although harmonization of manual hippocampal segmentation protocols has been initiated by several groups [3,4], such a tedious and error-prone procedure is not feasible in routine clinical practice. In this study, we aim at exploring the potential of an alternative automated method for hippocampal volumetry using T1-weighted images from the MP2RAGE sequence, which offer both low intensity bias and excellent grey-matter to white-matter contrast [5]. To validate its performance, we evaluate the ability of our volumetric measures to predict the Scheltens visual medial temporal atrophy (MTA) scores [6] on 27 patients undergoing clinical brain MRI for workup of cognitive decline.Material and methods
This study enrolled 27 patients (69% females, age range: 25-91 years, mean age: 67.6 years) who underwent clinical brain MRI for workup of cognitive decline. All patients were scanned at 3T (Magnetom Skyra, Siemens Healthcare, Erlangen, Germany) using the MP2RAGE sequence (TI1/TI2=700/2500ms, TR=5000ms, TA=8:22min, voxel size=1x1x1.2mm3 with a 64-channel head coil). Hippocampal atrophy severity was rated for each patient by two experts using the Scheltens 5-point rating scale (0-2: none to mild, 3-4: moderate to severe, see exemplary images in Figure 1).Automated segmentation of brain structures was performed with the MorphoBox [7] prototype which takes both the uniform MP2RAGE and the second inversion contrasts [5] as inputs. In a first step, the software estimates the total intracranial volume based on the product of the second inversion and uniform MP2RAGE images, as suggested in [8]. Next, a tissue classification is performed and hippocampus segmented using the uniform MP2RAGE image as described in [7]. The hippocampus volume is estimated by summing up gray- and white-matter a posteriori probabilities over left- and right-hippocampus masks obtained by atlas propagation [7]. Finally, for the purpose of inter-subject comparison, all estimated volumes are normalized by the total intracranial volume.
Multinomial logistic regression was used to study the correlation between automated hippocampus volume estimates and Scheltens scores averaged across the two raters. Classification accuracy, defined as the percentage of correct none-to-mild or moderate-to-severe hippocampal atrophy classification, was evaluated using leave-one-out cross-validation.
Results
The hippocampus volume estimates were found to be significantly correlated with the averaged Scheltens scores (p<0.0001). As seen in Figure 2, the median absolute prediction error, i.e. the difference between the averaged Scheltens scores and their predicted values using logistic regression, was 0.45, with six cases out of 27 yielding an error larger than one, and only one yielding an error larger than two. The estimated hippocampus volume turned out to be a particularly accurate predictor of whether or not the Scheltens score was larger than two, see Figure 3. Using the multinomial logistic regression model as a binary classifier of hippocampal volumetry according to none-to-mild or moderate-to-severe, the classification accuracy was estimated to be 96.3% using leave-one-out cross-validation.Discussion
Our results exhibit strong correlation between qualitative Scheltens ratings and quantitative hippocampal volumetry which is consistent with previous studies [1,2,3]. These findings are encouraging and indicate that an objective measure of hippocampal atrophy can be predicted with relatively high accuracy. Despite the low number of subjects enrolled in this study, our results suggest that the employed MP2RAGE-based hippocampal volumetry tool can integrate various spatial patterns of atrophy (e.g. widening of choroid fissure, widening of temporal horn or of the lateral ventricle, or decrease in hippocampal height) to a single score. This suggests that the method can be a biomarker of neurodegeneration on an individual patient basis.Acknowledgements
No acknowledgement found.References
1. Dubois, B, Feldman, HH, et al. Revising the definition of Alzheimer’s disease: a new lexicon. Lancet Neurol. 2010;11:1118-11272. Albert, MS, Dekosky, ST, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:270-279
3. Jack, CR, Barkhof, F, et al. Steps to standardization and validation of hippocampal volumetry as a biomarker in clinical trials and diagnostic criterion for Alzheimer’s disease. Alzheimers Dement. 2011;7:474-485
4. Frisoni, GB, Jack CR. Harmonization of magnetic resonance-based manual hippocampal segmentation: a mandatory step for wide clinical use. Alzheimers Dement. 2011;7:171-174
5. Marques, J. P., Kober, T., Krueger, G., van der Zwaag, W., Van de Moortele, P. F., & Gruetter, R. (2010). MP2RAGE, a self bias-field corrected sequence for improved segmen-tation and T 1-mapping at high field. Neuroimage, 49(2), 1271-1281.
6. Scheltens, P, Leys, D, et al. Atrophy of medial temporal lobes on MRI in « probable » Alzheimer’s disease and normal ageing : diagnostic value and neuropsychological correlates. J Neurol Neurosurg Psychiatry 1992;55:967-972
7. Schmitter, D, Roche A, et alAn evaluation of volume-based morphometry for prediction of mild cognitive impairment and Alzheimer's disease. NeuroImage: Clinical 2015;7:7-17.
8. Fujimoto, K, Polimeni, JR, et al. Quantitative comparison of cortical surface reconstructions from MP2RAGE and multi-echo MPRAGE data at 3 and 7T. Neuroimage 2014;90:60-73
Figures
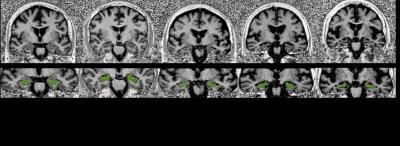
Figure 1. Exemplary coronal slices (top) of MP2RAGE uniform contrast going through the corpus of the hippocampus (level of the anterior pons) and corresponding hippocampus segmentation masks overlaid in green (bottom) of 5 randomly-picked patients with Scheltens scores spread over the 5-point rating scale
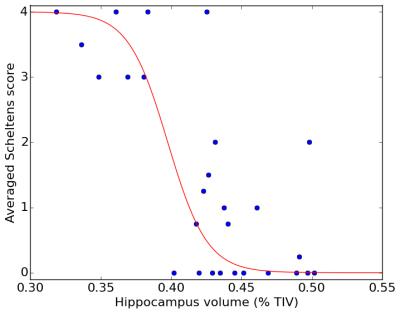
Figure 2. Illustration of multinomial logistic regression of average Scheltens scores in terms of normalized hippocampus volumes estimate by the MorphoBox prototype
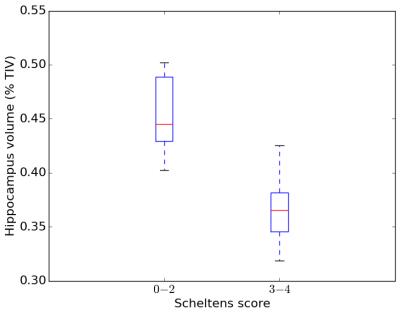
Figure 3. Distribution of estimated normalized hippocampus volumes depending on Scheltens-based atrophy severity (0-2: none to mild, 3-4: moderate to severe)