4116
Longitudinal Hypergraph Learning: A Consistent Segmentation Method for Measuring the Growth Trajectory of Infant Hippocampus from Brain MR Images1Department of Radiology and BRIC, The University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 2School of Computer and Information, Hefei University of Technology, Hefei, People's Republic of China
Synopsis
Automatic and consistent hippocampus segmentation from longitudinal infant brain MR image sequences is crucial for the measurement and analysis of its growth trajectory during early brain developing stage. To achieve this goal, we propose to use the longitudinal hypergraph method for joint learning the MR images from multiple acquisition time-points. We apply the proposed method to segment hippocampus from infant brain MR dataset which contains five time-points from 2 weeks to 12 months of age. According to the experimental results, our method outperforms other state-of-the-art label fusion methods in terms of both segmentation accuracy and consistency.
Introduction
During the first year of life, the infant brain undergoes a dramatic change in its physical and functional development. The hippocampus, located in the medial temporal lobe of the brain, has been discovered as an important brain structure serving for the spatial navigation and the short/long term memory. Therefore, the ability to accurately measure and analyze the growth trajectory of hippocampus from either normal subjects or subjects with neurological disorders is a crucial step in imaging-based early brain development studies. In this way, imaging biomarkers related to the hippocampus with disease can be further investigated for the clinical diagnosis and treatment plan. However, due to the myelination in the early stage of hippocampal formation, dramatic MR image contrast and appearance changes, as well as dynamic hippocampus shape variations, it is quite challenging to segment the infant hippocampus along multiple time-points. Therefore, the advanced technique is in great need to handle these challenges and to achieve both accurate and consistent infant hippocampus segmentation across multiple time-points.Methods
To overcome the inconsistency of hippocampus segmentations along multiple time-points, we propose to solve the multiple segmentation tasks along with all time-points as a single task, instead of processing each time-point independently like the conventional label fusion methods [1]. To achieve this goal, based on our previous work [2], a longitudinal hypergraph is built to model both the spatial voxel correlations within each time-point and also the temporal voxel coherence between each adjacent time-point pair. Specifically, two types of hyperedges are constructed to separately encode the spatial and temporal hippocampus neighborhood information. In this way, the proposed hypergraph is inherently adapted to solve the problem in segmenting the developmental hippocampus with dramatic appearance and shape changes. Furthermore, in order to take the advantage of multi-atlas label fusion method, we use a third type of hyperedge to build the relationships between the atlas images and the target image. In this way, the hypergraph model can make use of the group priors from atlas images for learning the labels on the target image. By enriching the hypergraph model with the above three types of hyperedges, the task of longitudinal segmentation can be formulated as a semi-supervised label fusion model. Thus, the label of each target voxel can be jointly and consistently determined not only from the labels of spatially and temporally neighboring voxels but also from the labels of atlas images.Results
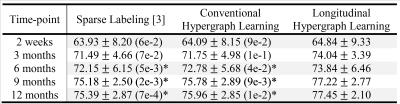
To evaluate the effectiveness of the proposed method, the dataset includes ten infant subjects. Each subject has the MR images scanned at 2 weeks, 3 months, 6 months, 9 months and 12 months of age, as well as their corresponding manual segmentations served as ground truth. For the comparison of segmentation accuracy and consistency with our method, sparse labeling [3] and conventional hypergraph learning which segments each time-point image independently are adopted. The leave-one-subject-out strategy is used to evaluate all the comparison segmentation methods.
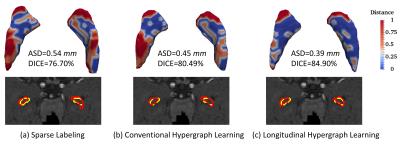
Table 1 shows the results of average Dice ratio (DICE) with standard deviation for three comparison segmentation methods at five time-points. It can be seen that our proposed method outperforms other two methods, especially for the last three time-points, which demonstrates superior segmentation accuracy and effectiveness of our method. Fig. 1 further visualizes the segmentation results of sparse labeling, conventional and longitudinal hypergraph learning methods for a typical subject. The first row shows their respective 3D surface distance maps to the ground-truth, along with the average surface distances (ASDs) and DICEs. The second row compares all the automatic segmentation contours with the ground truth contours. It can be seen that the highest similarity/accuracy can be found in longitudinal hypergraph learning method. Finally, to demonstrate the effectiveness of incorporating longitudinal information into the hypergraph model, we measure the volume ratio between the automatic segmentation and their ground truth. For a typical subject at 2-week, 3-month, 6-month, 9-month and 12-month, the volume ratios are 1.06, 1.27, 1.22, 1.13 and 1.65 by conventional hypergraph learning, and 0.98, 1.04, 1.10, 1.01, and 1.35 by longitudinal hypergraph learning, respectively. It can be observed that after introducing longitudinal constraint into the hypergraph, all ratios from five time-points are much closer to 1, which demonstrates better temporal consistency.
Conclusion
We propose a longitudinal hypergraph learning method for accurate and consistent segmentation of infant hippocampus from brain MR image across multiple time-points. Experimental results demonstrate that our proposed method is able to maintain both good temporal consistency along different time-points and high segmentation accuracy, which shows the feasibility of measuring and analyzing hippocampus growth trajectory for early brain developmental studies.Acknowledgements
This work is supported in part by National Institutes of Health (NIH) grants HD081467, EB006733, EB008374, EB009634, MH100217, AG041721, AG049371, AG042599, CA140413, MH088520 .References
1. Pipitone, J., et al.: Multi-atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. NeuroImage 2014; 101, 494-512.
2. Dong, P., Guo, Y., Shen, D., Wu, G.: Multi-atlas and Multi-modal Hippocampus Segmentation for Infant MR Brain Images by Propagating Anatomical Labels on Hypergraph. In: Wu, G., Coupé, P., Zhan, Y., Munsell, B., Rueckert, D. (eds.) First International Workshop, Patch-MI 2015, pp. 188-196. Springer; 2015
3. Wu, G., et al.: A generative probability model of joint label fusion for multi-atlas based brain segmentation. Medical Image Analysis 2014; 18, 881-890.
Figures