4094
The development of automatic 3D fetal brain MRI analysis methods for depicting growth trajectories of fetal brain tissues1Department of Imaging and Interventional Radiology, The Chinese University of Hong Kong, Hong Kong, Hong Kong, 2Shenzhen Research Institute,The Chinese University of Hong Kong, Shenzhen, People's Republic of China, 3Chow Yuk Ho Technology Center for Innovative Medicine, Hong Kong, 4Therese Pei Fong Chow Research Centre for Prevention of Dementia, 5Department of Medicine and Therapeutics, The Chinese University of Hong Kong, Hong Kong, 6Department of Radiology, Nanjing Drum Tower Hospital,The Affiliated Hospital of Nanjing University Medical School, Nanjing, People's Republic of China, 7Philips Healthcare, Hong Kong, 8Shenzhen Research Institute, The Chinese University of Hong Kong, Shenzhen, People's Republic of China
Synopsis
In this paper, we proposed a set of fetal brain MRI analysis methods to quantify the fetal brain tissue volume. We used deep learning-based brain mask extraction method to obtain brain mask and reconstructed 3D fetal brain volumes using registration-based reconstruction method. Then an age-specific atlas-based segmentation method was applied to segment three major tissues (White Matter, cortical Gray Matter, cerebrospinal fluid). The changes of intracranial volume and the three brain tissue volumes across different gestational ages were calculated and fitted with both linear and quadratic curves. The results demonstrated the effectiveness of the automatic 3D fetal MRI quantification methods.
PURPOSE
In order to understand how human brain develops in the early stage of life and meet the real clinical demand in early identification of fetal pathologies, fetal magnetic resonance imaging (MRI) has emerged as an important clinical research and diagnostic tool in prenatal medicine because of its superior soft tissue contrast compared with ultrasound images. However, due to the research on 3D fetal MRI quantification still lags behind compared with adult MRI analysis, most of the current fetal MRI diagnosis and research still focus on 2D, with a limited number of exploratory 3D fetal MRI studies. In this study, we propose a set of image processing and analysis methods to reconstruct and analyze 3D fetal brain MRI volume, and perform a preliminary study on quantifying the brain tissue development in prenatal stage.DATA
We have collected T2-weighed MRI images of 30 healthy fetuses from 24 to 37.6 gestational weeks (GW) (29.6±3.9 GW) from Nanjing Drum Tower Hospital, China. All of the fetal MRI used were read as normal by a neuroradiologist. The fetal MRIs were acquired on a Philips Archieva 1.5T machine with eight-element abdominal phased array coil using SSFSE sequence in 3 nominally orthogonal anatomical planes, with axial direction perpendicular to the brain stem. Three image stacks are acquired for each plane. TR=4500ms, TE=91ms, flip angle=90°, in-plane matrix size=400×400, FOV=300×300mm, slice thickness/gap=3/0mm with interleaved acquisition, slice number=30.METHODS
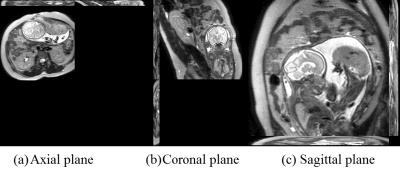
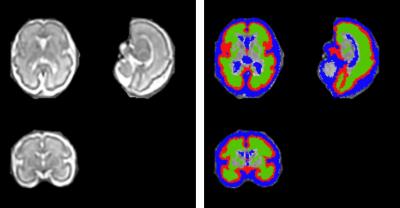
With the acquired multiply 2D MRI stacks for each subject (one 2D MRI example shown in Fig.1), a new deep learning model-based 2D brain extraction method was applied to extract fetal brain mask slice-by-slice [1]. With the brain mask, we applied the slice-to-volume registration based super-resolution reconstruction method to reconstruct the 3D volume and in the meanwhile correct the motion artifacts and bias field [2]. Then we normalized the 3D fetal brain MRI with age-specific fetal brain atlas [3] using rigid registration. Symmetric normalization (SyN) non-rigid registration [4] was performed to transform the tissue labels in the atlas space to the subject space and obtain the tissue labels (i.e., white matter (WM), cortical grey matter (cGM) and cerebrospinal fluid (CSF)) for each subject. The 3D reconstructed fetal MRI (the same subject as in Fig.1) and its segmentation results are shown in Fig. 2.RESULTS
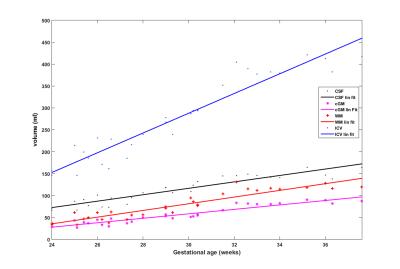
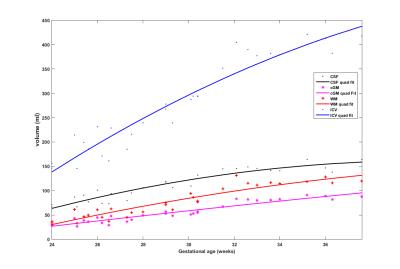
We measured the intracranial volume (ICV) and three brain tissue volumes for each subject and fit the volumes with the gestational age of the subject. Fig. 3 and 4 shows the growth trajectory of ICV, volumes of WM, cGM and CSF. Both linear fit (Fig.3) and quadratic fit (Fig.4) were performed to fit each set of samples. It can be observed that ICV and volumes of all the three brain tissues are increasing with gestational age. We used R2 to measure goodness of fit of each curve: ICV (linear: 0.9058; quad: 0.9138), WM (linear: 0.8660; quad: 0.8746), cGM (linear: 0.8984; quad: 0.8989), CSF (linear: 0.8647; quad: 0.8934). For all the curves, quadratic fit shows a slight better fitness compared to the linear fit.DICUSSION AND CONCLUSION
In this paper, we developed a set of methods to perform 3D fetal brain MRI analysis. First of all, we extracted brain mask automatically using deep learning model and reconstructed high quality 3D volumes from multiple stacks of 2D MRIs. Then with a 4D age-specific fetal brain atlas, we performed atlas-based segmentation to segment several major brain tissues. The growth trajectories of these brain tissues and whole brain were measured. In this preliminary study, we use the current data to verify the effectiveness of the proposed 3D fetal MRI analysis methods. With the continuously accumulated fetal brain MRI data, we will measure the volumetry of the fetal brain tissues across different gestational ages in the future.Acknowledgements
No acknowledgement found.References
[1] Li J, Luo Y, Shi L, Zhang X, Zhang B, Wang D. Automatic fetal brain extraction from MRI using deep learning for accurate 3D brain reconstruction. IEEE Transactions on BME under review.
[2] Kainz B, Steinberger M, Wein W, Murgasova M, Malamateniou C, Keraudren K, Aljabar P, Rutherford M, Hajnal J, Rueckert D. Fast Volume Reconstruction from Motion Corrupted Stacks of 2D Slices. IEEE Transactions on Medical Imaging 2015,34(9):1901-13.
[3] A Gholipour, C Limperopoulos, S Clancy, C Clouchoux, A Akhondi-Asl, J A Estroff, and S K Warfield. Construction of a Deformable Spatiotemporal MRI Atlas of the Fetal Brain: Evaluation of Similarity Metrics and Deformation Models. MICCAI, 2014, 17(0 2): 292–299.
[4] Avants, B. B., Tustison, N. J., Song, G., Cook, P. A., Klein, A., Gee, J. C. (2011). A Reproducible Evaluation of ANTs Similarity Metric Performance in Brain Image Registration.NeuroImage, 2011, 54(3),2033-2044.
Figures