4086
Mapping and modulation of Down Syndrome specific functional network in Dp(16)1yey mouse model1Engineering science, computer science and imaging laboratory (ICube), University of Strasbourg-CNRS France, Strasbourg, France, 2Biophysics and Nuclear Medicine, Faculty of Medicine and University Hospital, Strasbourg, Strasbourg, France, 3Medical Physics, AMIR, University Hospital, Freiburg, Freiburg, GA, Germany, 4Translational medicine and neurogenetics, Institute of Genetics and Molecular Imaging, University of Strasbourg
Synopsis
Resting state functional MRI (rsfMRI) is currently the only non-invasive approach capable of giving insight into the large-scale cerebral networks architecture and its dynamic changes in pathology or following therapeutic interventions. With the aim of deciphering specific network signatures underlying memory and cognitive impairments in Down Syndrome pathology, we performed rsfMRI and network analysis in the Dp(16)1yey mouse model. We found perturbed synchrony of BOLD-signal in the hippocampal network of Dp(16)1yey mice. We further modulated this memory specific cerebral circuitry via therapeutic treatment with a DYRK1A kinase inhibitor, aimed at rescuing the memory and cognitive dysfunctions characterizing this mouse model.
Introduction and Aims:
Down syndrome (DS) is characterized by the presence of an extra copy of human chromosome 21 (HSA21) and represents the most common genetic cause for developmental cognitive disability in humans1,2. DS patients have low IQ values, memory impairments as well as difficulties in acquiring new skills3. One of the chromosome 21 genes, the serine/threonine kinase DYRK1A gene, was previously proposed as candidate for inducing the observed cognitive deficits5,6. Indeed, over-dosage of DYRK1A was found to correlate with memory/cognitive impairments in mice7,8. Despite the substantial progress in the understanding of the effect of gene overexpression at the molecular level, there is a paucity of information on the consequences on overall brain functional organization and connectivity. RsfMRI breaks new ground in neuroscience offering a non-invasive window into the intrinsic organization of functional cerebral networks9,10 and addressing the issue of pathology to network or treatment to network signatures.In this context, our study had two main objectives: (i) to characterize the functional connectional architecture in the Dp(16)1yey mouse model of DS via rsfMRI and to identify vulnerable networks; and (ii) decipher – whether and how - the functional connectivity is restored or remodeled upon therapeutic treatment aimed at reversing memory and cognition deficits.Materials and methods
Animal model.
Dp(16)1yey mouse is characterized by a duplication of a 22,9Mb region on mouse MMU16 chromosome. and over-express the serine/threonine kinase DYRK1A11,12. The mice show affected memory and cognitive skills. The model represents the ideal tool to test specific DYRK1A kinase inhibitors, such as L4113 as treatment of cognitive DS phenotype.
Experimental set-up: 3 months old Wild Type (WT) and Dp(16)1yey adult mice were included in this study (Groups 1 et 2: WT and Dp(16)1yey – Saline treated; Groups 3 et 4: WT and Dp(16)1yey - L41 treated). L41 was administrated via daily IP injection for 19 days in animal groups 3 and 4, as previously reported15. In the same cohort of animals the novel objects recognition (NOR) task16 tested memory performance.
MRI experiments were carried-out at the end of L41 or Saline treatment: Mouse physiological conditions were monitored during imaging performed under medetomidine (MD) sedation. MRI was performed with a 7T small bore animal scanner (Biospec 70/20, Bruker, Germany) and a mouse head adapted cryocoil. rsfMRI data was acquired using single shot GE-EPI (TE/TR=10ms/1700ms), 20 axial slices of 0.7mm thickness (FOV:19.2×12mm², resolution:0.15×0.15mm2, 400 volumes).
Data analysis: rsfMRI data was pre-processed using SPM8 for motion correction, spatial normalization with a study based template and the Allen Mouse Brain Atlas space. This allowed atlas based generation of regions of interest (ROIs) further used as nodes to compute the group-specific mouse brain connectivity matrices. These ROIs were also used in a seed-based correlation analysis.
Results and discussion:
Dp(16)1yey mouse model is characterized by perturbed intrinsic functional connectivity. We exemplify here the results obtained for hippocampus (Fig 1.), a brain area generally associated with performance in the NOR tasks. Disrupted synchrony of the BOLD signal is observed in the Dp(16)1yey hippocampal network, with spatially restrained patterns compared to the wild type group (Fig 1, A and B). Lower correlation coefficients are quantified within the hippocampal area as well, highlighting general under-connectivity fingerprints for this specific – memory related network. This modification is correlated with significant memory/cognitive deficits assessed in the Dp(16)1yey mice included in this study (Fig 2). We further performed whole brain network analysis using graph theory and found altered “hubs” networks configurations in Dp(16)1yey model. Specific inhibition of the DYRK1A kinase activity with L41 resulted in remodeling of the hippocampal FC pattern (Fig 1, C, D). L41 acted on both wild type (Fig 1 C) and Dp(16)1yey brains. However, L41 promoted stronger coherence of BOLD fluctuations in hippocampus of DS model, suggesting possible rescuing of the memory and cognitive impairements, as previously observed14.
Conclusion
We identified FC alterations that are specific to DS pathology. We show that hippocampal network strongly react to therapeutic modulations in the Dp(16)1yey brains. Further analysis will unravel the impact of L41 treatment at large scale network level. The results will be compared with analysis of DTI data acquired in the same imaging session. This aproach would provide a full picture of connectome modifications in Dp(16)1yey pathologic brains and after therapy.Acknowledgements
No acknowledgement found.References
1. Hook, E.G. (1982) Epidemiology of Down syndrome. In Pueschel, S.M. and Rynders, J.E. (eds), Down Syndrome. Advances in Biomedicine and the Behavioral Sciences, Ware Press, Cambridge, p. 11. 2. Pennington, B.F., Moon, J., Edgin, J., Stedron, J. and Nadel, L. (2003) The neuropsychology of Down syndrome: evidence for hippocampal dysfunction. Child Dev., 74, 75–93. 5.
3. Pulsifer, M.B. (1996). The neuropsychology of mental retardation. J. Int. Neuropsych. Soc., 2, 159–176.
4. Chapman, R.S. and Hesketh, L.J. (2000) Behavioral phenotype of individuals with Down syndrome. Ment. Retard. Dev. Disabil. Res. Rev., 6, 84–95.
5. Garcia-Cerro, S., Martinez, P., Vidal, V., Corrales, A., Florez, J., Vidal, R., Rueda, N., Arbones, M.L., and Martinez-Cue, C. (2014). Overexpression of Dyrk1A is implicated in several cognitive, electrophysiological and neuromorphological alterations found in a mouse model of Down syndrome. PloS one 9, e106572.
6. Jiang, X., Liu, C., Yu, T., Zhang, L., Meng, K., Xing, Z., Belichenko, P.V., Kleschevnikov, A.M., Pao, A., Peresie, J., et al. (2015). Genetic dissection of the Down syndrome critical region. Hum Mol Genet.
7. Souchet, B., Guedj, F., Penke-Verdier, Z., Daubigney, F., Duchon, A., Herault, Y., Bizot, J.C., Janel, N., Créau, N., Delatour, B., et al. (2015). Pharmacological correction of excitation/inhibition imbalance in Down syndrome mouse models. Front Behav Neurosci 9, 267.
8. Guedj, F., Pereira, P.L., Najas, S., Barallobre, M.J., Chabert, C., Souchet, B., Sebrie, C., Verney, C., Herault, Y., Arbones, M., et al. (2012). DYRK1A: a master regulatory protein controlling brain growth. Neurobiology of disease 46, 190-203.
9. Leergaard TB, Hilgetag CC, Sporns O. (2012). Mapping the connectome: multi-level analysis of brain connectivity. Front Neuroinform. May 1;6:14
10. van den Heuvel MP1, Hulshoff Pol HE. (2010). Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur Neuropsychopharmacol. 2010 Aug;20(8):519-34.
11. Li Z, Yu T, Morishima M, Pao A, LaDuca J, Conroy J, Nowak N, Matsui S, Shiraishi I, Yu YE.(2007). Duplication of the entire 22.9 Mb human chromosome 21 syntenic region on mouse chromosome 16 causes cardiovascular and gastrointestinal abnormalities. Hum Mol Genet. 2007 Jun 1;16(11):1359-66.
12. Souchet, B., Guedj, F., Sahul, I., Duchon, A., Daubigney, F., Badel, A., Yanagawa, Y., Barallobre, M.J., Dierssen, M., Yu, E., et al. (2014). Excitation/inhibition balance and learning are modified by Dyrk1a gene dosage. Neurobiology of Disease Neurobiol Dis. Sep;69:65-75.
13. Tahtouh, T., Elkins, J.M., Filippakopoulos, P., Soundararajan, M., Burgy, G., Durieu, E., Cochet, C., Schmid, R.S., Lo, D.C., Delhommel, F., et al. (2012). Selectivity, cocrystal structures, and neuroprotective properties of leucettines, a family of protein kinase inhibitors derived from the marine sponge alkaloid leucettamine B. J Med Chem 55, 9312-9330.
14. Guedj, F., Sebrie, C., Rivals, I., Ledru, A., Paly, E., Bizot, J.C., Smith, D., Rubin, E., Gillet, B., Arbones, M., et al. (2009). Green tea polyphenols rescue of brain defects induced by overexpression of DYRK1A. PloS one 4, e4606.
15. Reeves, R.H., Irving, N.G., Moran, T.H., Wohn, A., Kitt, C., Sisodia, S.S., Schmidt, C., Bronson, R.T., and Davisson, M.T. (1995). A mouse model for Down syndrome exhibits learning and behaviour deficits. Nature genetics 11, 177-184.
16. Bevins RA1, Besheer J. (2006) Object recognition in rats and mice: a one-trial non-matching-to-sample learning task to study 'recognition memory'. Nat Protoc. 2006;1(3):1306-11.
Figures
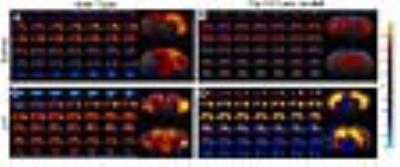
Hippocampal functional network as mapped via seed correlation in the four experimental groups: WT and Dp(16)1yey – Saline treated (A and B); WT and Dp(16)1yey - L41 treated (C and D). The hippocampal ROI was derived from the Allen Mouse Brain Atlas.
Dp(16)1yey mice showed disrupted synchrony of the BOLD signal (B) in the hippocampal network, with spatially restrained patterns compared to the WTgroup.
Specific inhibition of the DYRK1A kinase activity with L41 resulted in remodeling of the hippocampal FC pattern. L41 acted on both WT (C) and Dp(16)1yey brains (D). L41 promoted stronger FC within hippocampus of DS model,
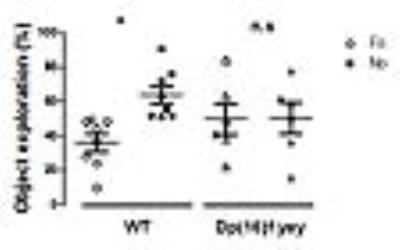
Memory performance in WT and Dp(16)1yey mice assessed via novel object recognition (NOR) test. Significant memory/cognition impairments are noticed in Dp(16)1yey mice.
NOR test is based on the innate tendency of rodents to differentially explore novel objects (NO) over familiar (FO) ones. After an acquisition phase, mice returned to their cage for a 24 hours retention interval. To test their memory, on day 3, one FO and one NO were placed in the apparatus and mice were free to explore. Object exploration was manually scored. The percent of time exploring familiar vs novel objects was calculated to assess memory performance.