4064
ACCURACY AND REPRODUCIBILITY OF AN AUTOMATED LESION SEGMENTATION TOOL-LESIONQUANT1CorTechs Labs, San Diego, CA, United States, 2Institute of Biomedical Engineering, Bogazici University, Istanbul, Turkey
Synopsis
Quantitative measures such as lesion volume and distribution have significant value for clinicians evaluating disease progression. Clinical standards for lesion evaluation include visual inspection of MRI images, or expert manual segmentation of lesions. These subjective measurements are often vulnerable to inter- and intra-rater variability, resulting in low reproducibility. CorTechs Labs’ LesionQuant is a fully-automated lesion segmentation tool for clinical use designed to provide accurate and reproducible lesion segmentations. This study objectively evaluates the segmentation results of LesionQuant compared to expert manual segmentation.
PURPOSE
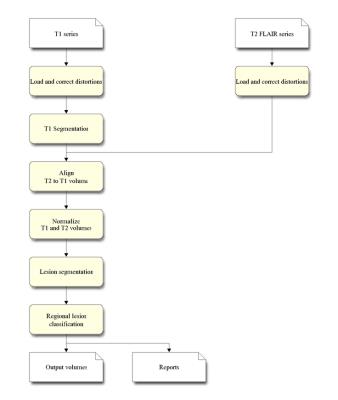
Quantitative measurements of lesion volume and distribution have significant value for clinicians evaluating disease progression. Subjective measurements based on clinician’s visual inspection and manual segmentation are often vulnerable to inter- and intra-rater variability, resulting in low reproducibility. Thus, objective automated lesion segmentation tools have been developed to overcome these problems. Since the appearance of lesions varies across MRI protocols, incorporating different MRI scans provides more information for lesion segmentation tools when delineating lesions. In current clinical practice, a 3D T1-weighted scan acquired with 1.2 mm slices is recommended for volumetric analysis of global measures and change of important structures over time. T2 FLAIR imaging is recommended for evaluating brain lesions since T2 FLAIR is the most sensitive measure of white matter lesion in the cerebrum. Therefore, combining the T1-weighted and T2 FLAIR data could result in the optimal lesion segmentation. LesionQuant, a NeuroQuant product (CorTechs Labs Inc., San Diego, California, USA), is a fully-automated lesion segmentation tool for clinical use. It was designed to provide accurate and reproducible lesion segmentations using both T1-weighted and T2 FLAIR data. In this study, objective methods were implemented to evaluate LesionQuant’s lesion segmentation performance.METHODS
RESULTS
For the total lesion segmentation, the accuracy test showed Pearson’s correlation coefficient was 0.98, ICC was 0.94 and Dice coefficient was 83.4% ± 7.5%. Reproducibility analysis showed that Dice coefficient was 89.0% ± 3.5% and absolute difference was 1.0% ± 0.8%.DISCUSSION
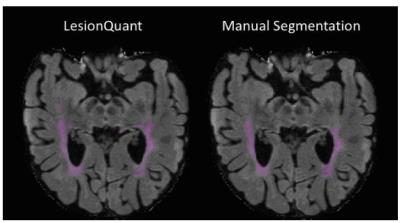
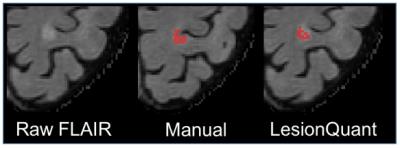
Initial results demonstrated good overall reproducibility for repeat scans. The accuracy results showed high agreement between LesionQuant and expert manual segmentation as shown in Figure 2 and Figure 3. There are several limitations to the analyses conducted in this initial study. The number of participants included in this study is relatively small when considering the variability of lesion presentation in MS and other white matter diseases. In addition, further studies should focus on the inclusion of additional MS participants in order to capture the full spectrum of lesion distributions and disease severity. Future analyses should also incorporate patients with a variety of diseases that can present with T2 FLAIR lesions. Future analyses will also compare LesionQuant software with automatic and semi-automatic research tools that are freely available for lesion segmentation.CONCLUSION
The initial results of this study demonstrate that the accuracy of LesionQuant is comparable to expert manual segmentation of lesions and had high reproducibility. This suggests that LesionQuant could be a useful tool in quantitative analysis of the brain lesions.Acknowledgements
No acknowledgement found.References
1. Surya Probha D. Satheesh Kumar J. Performance Evaluation of image Segmentation Using Objective Methods, Indian J. of Science and Technology, Vol 9(8), 2016
2. Zhang YJ., Evaluation and comparison of different segmentation algorithms, Pattern Recognition, 1996; 29(8):1335-46
3. Zhang H., Fritts, JE, Goldman SA, Image segmentation evaluation: A Survey of Unsupervised methods.
4. Pustian D, etc., Automated Segmentation of Chronic Stroke Lesions Using LINDA: Lesion identification with neighborhood data analysis, Hum Brain Mapp. Apr;37(4):1405-21, 2016
5. Sahraian, A.S et al, MS Lesions in Fluid Attenuated Inversion Recovery Images. MRI Atlas of MS Lesions. pg. 35-44, 2008, Springer, Berlin, Heidelberg