4001
Unstreaking: Radial MRI with Automatic Streaking Artifact ReductionLi Feng1, Hersh Chandarana1, Daniel K Sodickson1, and Ricardo Otazo1
1Center for Advanced Imaging Innovation and Research (CAI2R), New York University School of Medicine, New York, NY, United States
Synopsis
Streaking artifact is one of the major causes of image quality degradation in radial MRI. Since multicoil arrays are widely used in modern MR scanners, an easy way to reduce streaking artifacts is to identify coil elements that are contaminated by a high level of streaks, and then exclude them from image reconstruction. However, such an approach requires accurate clustering algorithms to automatically select unwanted coil elements. In this work, a method called “Unstreaking” is proposed for automatic streaking artifact reduction without the need to exclude coil elements. The method was tested for accelerated radial DCE-MRI of the liver.
INTRODUCTION
Streaking artifact is one of the major causes of image quality degradation in radial MRI1. Although streaks normally arise from the edge of the field of view (FOV) due to off-resonance, gradient nonlinearity and/or insufficient fat suppression, they often spread throughout the entire image. Multiple receiver coils with different spatial sensitivities can be used to localize the source of these artifacts, and several approaches have been proposed to exclude coil elements containing a high level of streaking artifacts2,3. These coil elements are usually located far from the center of the FOV and often contribute little to image information. However, a major challenge of such approaches is that an accurate clustering algorithm is required to automatically select unwanted coils. Incorrect clustering may lead to loss of important image information or insufficient removal of streaks. In this work, we propose a method to reduce streaking artifact in radial imaging without the need for excluding coil elements. The method first calculates the streaking artifact level of each coil element, and then adjusts the contribution of each coil to the final image by weighing the corresponding k-space data accordingly. The proposed method was tested for accelerated radial DCE-MRI of the liver.THEORY
The concept of streak ratio was proposed by Xue Y et al2 for evaluating the degree of streaking artifacts. In this study, we define our streak ratio in a different way, as shown in Figure1. Here, Ref refers to multicoil images reconstructed using a number of spokes (e.g., 800) that is sufficient to satisfy the Nyquist criterion; Img indicates multicoil images reconstructed using a small number of spokes (e.g., 40), in which a high level of artifact is generated due to heavy undersampling; Diff is the difference of Ref and Img for quantifying the artifact level created in each coil. Since undersampling artifacts in radial imaging are dominated by streaks, the streak ratio for each coil can be calculated using Equation 1 shown in Figure1. In a second step, instead of clustering coils and excluding those contaminated by strong streaking artifacts, our method aims to adjust the intensity of k-space in each coil element based on its streak ratio, so that their contribution to the final image is weighted accordingly (i.e., coil elements with a high streak ratio contribute less to the result, and vice versa). This process is called “Unstreaking,” and a detailed pipeline is outlined in Figure1. The exponent a in Equation 2 controls the degree of penalty for each coil. A higher value of a leads to stronger penalization of streaking artifacts but can also cause increase in signal inhomogeneity due to excessive weighting. As shown in Figure1, Unstreaking can clearly reduce the streak level for a =1.METHODS
IRB-approved liver DCE-MRI was performed in three volunteers in a transverse orientation using a golden-angle stack-of-stars sequence. Imaging protocols were prescribed similar to that proposed in (4), and data were continuously acquired for 190 seconds. Post-contrast liver imaging was also performed in a patient in a coronal orientation after all clinical scans using the same imaging protocol, and data were continuously acquired for 90 seconds. DCE liver datasets were reconstructed using the GRASP (Golden-angle RAdial Sparse Parallel) technique described in (4) both with and without Unstreaking. The post-contrast liver dataset was reconstructed using XD-GRASP (eXtra-Dimensional GRASP)5 in a similar manner.RESULTS
Figure2 shows GRASP (top) and GRASP-Unstreaking (bottom) results from the first volunteer. As shown by the green arrows, GRASP results suffered from streaking artifacts that originated from some regions (e.g., fat signal in the arms) outside the FOV. GRASP-Unstreaking, on the other hand, improved image quality in different contrast-phases with reduced the streaking artifacts, at the cost of a slight increase in signal inhomogeneity. GRASP-Unstreaking achieved similar improved performance in all cases, as shown in Figure3 and Figure4. Figure5 shows results with (bottom) and without (top) Unstreaking in the coronal plane. In this subject, substantial streaking artifacts were generated at the edge of FOV in both NUFFT and XD-GRASP results, while Unstreaking significantly improved image quality, with highly-reduced streaking artifacts.DISCUSSION
An approach called Unstreaking is proposed in this study for automatic streaking artifact reduction in radial MRI. Compared to other techniques that require an algorithm to exclude coil-elements contaminated by streaks, Unstreaking aims to weight k-space according to the streak ratio calculated for each coil. Since Unstreaking is a pre-reconstruction step, it does not interfere with acceleration techniques such as compressed sensing and/or parallel imaging. Our initial results suggest that Unstreaking can be useful for clinical application of radial sampling.Acknowledgements
This work was supported in part by the NIH, and was performed under the rubric of the Center for Advanced Imaging Innovation and Research (CAI2R), a NIBIB Biomedical Technology Resource Center (NIH P41 EB017183).References
[1] Du J et al. MRM 2004; 51:1071–1076 [2] Xue Y et al. MRM 2012; 67:470–476 [3] Grimm R et al. ISMRM 2013; p3786 [4] Feng L et al. MRM 2014; 72 (3), 707-717 [5] Feng L et al. MRM 2016; 75 (2), 775-788Figures

Figure 1: Unstreaking procedure: Ref is multicoil images reconstructed using a number of spokes
(e.g., 800) that is sufficient to satisfy the Nyquist criteria; Img is multicoil images reconstructed
using a small number of spokes (e.g., 40), in which a high level of artifacts
is generated due to heavy undersampling; Diff
is the differences of Ref and Img for quantifying the artifacts level
created in each coil-element. The intensity of each k-space is then adjusted
based on its streak ratio calculated using Equation 1, so that the contribution
of each k-space to the final image is weighted accordingly.
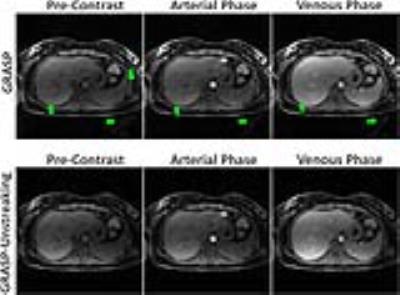
Figure2: Comparison of GRASP (top row) and
GRASP-Unstreaking (bottom row) results in one volunteer. GRASP results suffered
from streaking artifacts (green arrows) that originated from some regions (e.g.,
fat signal in the arms) outside the FOV due to off-resonance and/or gradient
nonlinearity effects. GRASP-Unstreaking, on the other hand, improved image
quality in different contrast-phases with reduced the streaking artifacts, at
the cost of slight increase of signal inhomogeneity.
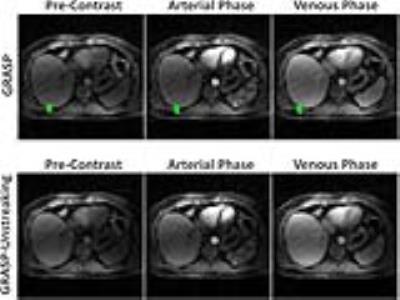
Figure3: GRASP-Unstreaking achieved better performance with
reduced streaking artifact comparing to standard GRASP reconstruction.
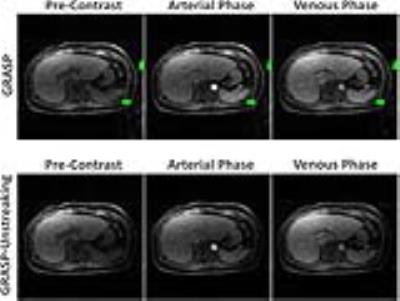
Figure 4: GRASP-Unstreaking achieved better performance
with reduced streaking artifact comparing to standard GRASP reconstruction.
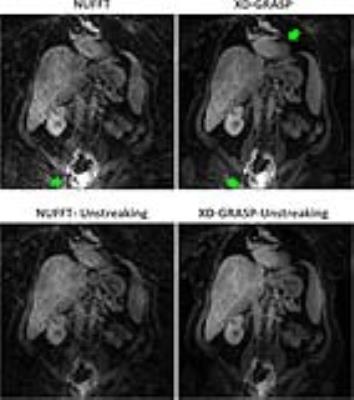
Figure 5: Comparison of coronal images with (bottom
column) and without (top column) Unstreaking. Substantial streaking artifacts
were generated at the edge of FOV in standard NUFFT and XD-GRASP results, while
Unstreaking significantly improved image quality with highly-reduced streaking
artifacts.