3996
A Novel Multi-Center Classification Method for ASD Diagnosis via Sparse Multi-Modality Multi-Task Learning1Department of Radiology and Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 2School of Digital Media, Jiangnan University, Wuxi, People's Republic of China, 3Med-X Research Institute, School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, People's Republic of China, 4Department of Biomedical Engineering, Faculty of Engineering, National University of Singapore, Singapore
Synopsis
Multi-task classification targeting multi-center ASD diagnosis is not well investigated yet. Taking advantages of the Autism Brain Imaging Data Exchange (ABIDE) database, we propose a novel multi-modality multi-center classification (M3CC) method for accurate ASD diagnosis. We formulate the diagnosis into a multi-task learning problem, as each task corresponds to the classification of the subjects of one center. Our comprehensive experiments show that, by incorporating multi-modality neuroimaging data and handling multiple centers jointly, the performance of computer-assisted ASD diagnosis is increased significantly.
Purpose
Most imaging-based ASD classifiers are limited to single imaging centers, which often struggle with datasets of limited sizes and thus suffer from poor ASD diagnosis capability. To this end, we develop a novel multi-task classification method, which attains ASD diagnosis based on multi-modality multi-center images. Higher diagnosis accuracy is achieved by comprehensively incorporating multi-modality data and jointly conducting multiple classification tasks of multiple centers.Methods
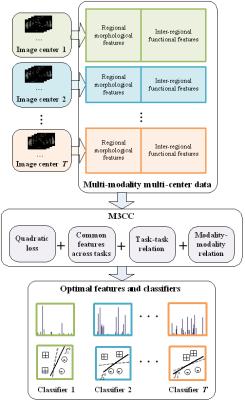
Autism spectrum disorder (ASD) is characterized as a syndrome of poor social communication abilities in combination with repetitive behaviors or restricted interest. Previous studies have indicated that multi-modality classifiers can achieve better performance than using a single modality in diagnosing ASD. However, most of them are limited to the datasets acquired from single imaging centers, which do not always provide enough training data for accurate ASD classification. In clinical practice, it is common to acquire images from multiple centers and handle them simultaneously. Thus, we develop a novel ASD diagnosis method for multi-modality and multi-center (M3CC) imaging data. The M3CC method, which is illustrated in Fig. 1, is featured by its capability of handling the inconsistency across the images of multiple centers.
In our study, we consider both T1-weighted MRI and resting-state functional MRI (rs-fMRI) scans acquired from multiple imaging centers. Regional morphological features are extracted from the T1-weighted MRI in an automated manner using the standard FreeSurfer pipeline. This process generates 303 regional morphological features, including the cerebral cortical gray matter (GM) volumes, subcortical white matter (WM) volumes, mean cortical thickness, the subcortical structure volumes, Brodmann area volumes and thickness. Meanwhile, we extract features from rs-fMRI using the pipeline provided by ABIDE with AFNI (https://afni.nimh.nih.gov/afni/). The Automated Anatomical Labeling (AAL) template is utilized in this process and a 116×116 functional connectivity matrix is generated for each subject. The correlation measures in the upper triangle of the matrix are treated as the inter-regional functional features. In this way, both regional morphological features and inter-regional functional features are extracted for subsequent multi-task learning.
We treat the classification of each imaging center as one task. The group sparsity as well as both task-task and modality-modality relations are devised as the regularization terms during learning. The group sparsity guides the joint selection of the common features across different tasks. Regarding the task-task relation, we assume that, if two centers are closely related, their corresponding classifiers should be similar. In modality-modality relation, we assume that the classifiers for different modalities of one subject tend to reach the same disease label. We develop an efficient iterative optimization solution to our formulated problem. In this way, the optimal feature selection and the modeling of classifiers can be jointly conducted for highly accurate ASD diagnosis.
Results
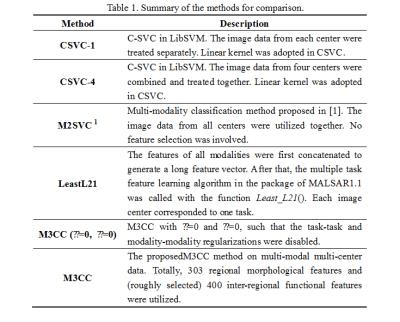
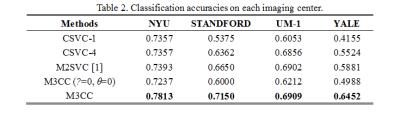
Our experiments are based on both T1-weighted MRI and rs-fMRI scans in ABIDE. All subjects included are under 15 years old and recruited from four different imaging centers, i.e., NYU, STANDFORD, UM_1 and YALE. A 10-fold nested cross-validation was adopted to evaluate the performance. Table 1 summarizes all methods under comparisons and Table 2 shows the comparison results, from which one may observe the superiority of M3CC over other methods.
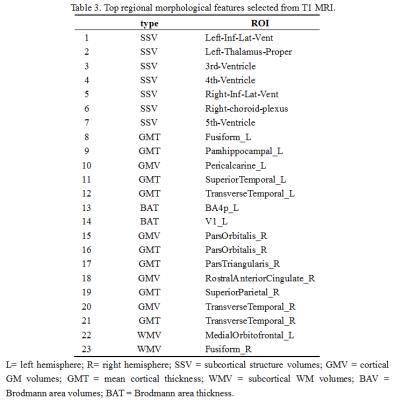
Table 3 shows the discriminative regional morphological features selected by M3CC. One can observe that the selected features included measures from subcortical structure volumes, mean cortical thickness, cortical GM volumes, and subcortical WM volumes. The diversity of the sources of discriminative features indicates that different types of regional morphological features were complementary to each other when identifying ASD from healthy controls. It is also observable that the selected features were distributed in various brain regions, which indicates the spread of morphological abnormalities over the whole brain in ASD patients.
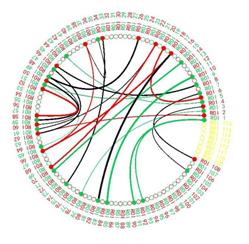
M3CC can also find the most discriminative functional connections for classification. Fig. 2 visualizes the common discriminative connections shared by four imaging centers using connectogram. One can observe that more intra-hemisphere connections existed in the left hemisphere than in the right, which is consistent with the left-hemisphere hypothesis for ASD2. Besides, the selected connections involved multiple cortical regions and subcortical structures related to ASD.
Acknowledgements
This work was supported in part by NIH grants (EB006733, EB008374, EB009634, MH100217, MH108914, AG041721, AG049371, AG042599, DE022676), and by the National Natural Science Foundation of China (61300151, 61401271, 61473190, 81471733), Science and Technology Commission of Shanghai Municipality (16511101100), the Natural Science Foundation of Jiangsu Province (BK20151299, BK20151358 and BK20161268).References
[1] Zhang D, Wang Y, Zhou L, et al. Multimodal classification of Alzheimer's disease and mild cognitive impairment[J]. Neuroimage, 2011, 55(3): 856-867
[2] Chandana S R, Behen M E, Juhász C, et al. Significance of abnormalities in developmental trajectory and asymmetry of cortical serotonin synthesis in autism[J]. International Journal of Developmental Neuroscience, 2005, 23(2): 171-182.
Figures