3989
Robust and Efficient Dictionary Matching in Magnetic Resonance Fingerprinting with Neural NetworksDaniel Truhn1, Christoph Haarburger2, Volkmar Schulz3, Dorit Merhof2, and Christiane Kuhl1
1Radiology, University Hospital Aachen, Aachen, Germany, 2Institute of Imaging and Computer Vision, 3Physics of Molecular Imaging Systems
Synopsis
We implemented efficient and robust matching of signals acquired in magnetic resonance fingerprinting by use of neural networks and show its superiority in terms of speed and robustness to noise.
Purpose
Magnetic resonance fingerprinting (MRF) is a novel technique that has recently gained attention due to its ability to rapidly acquire simultaneous multi-parametric quantification of tissue parameters. A key step in this approach is the matching of the signal amplitude of a specific measurement to a pre-calculated dictionary in order to extract the relevant tissue parameters (T1, T2, Δf etc.). This step proves to be computationally demanding and poses a serious problem. Methods to shorten computation times have been employed recently. Among the more promising methods are the use of principal component analysis and kd-tree search which reduce matching times by 2-3 orders of magnitude. However, matching time may still increase with noise levels and parameters such as the threshold value in PCA have to be tuned and adapted accordingly. Furthermore, computation time still poses a problem when dictionaries become large (e.g. matching of more than three parameters), image resolution becomes high or when iterative reconstruction schemes are employed. In this work we describe how deep neural networks can be efficiently trained and employed to the matching problem. We show that they are superior in matching speed, accuracy and storage requirements.Methods
The bloch equations were simulated for a gradient echo sequence with pseudorandom repetition times between 10 and 14 ms and a sinusoidal pattern of flip angles for 500 excitations (1). Our neural network consists of an encoding- and a regression part. The encoding part is composed of three layers incorporating 1024, 512 and 256 units respectively. This part of the network is pretrained as a denoising autoencoder which leads to an encoded noise-robust signal representation. During pretraining zero-mean Gaussian noise with std=0.15 from the maximum signal amplitude is added to the signal at the input. After pretraining, the encoding layers’ parameters are frozen and a 256 unit regression layer is added to learn the mapping between encoded representations and tissue parameters. Results of the neural network mapping are compared with a conventional dictionary matching algorithm. Computation times, robustness with respect to noise and precision were examined by analyzing signals with different amounts of noise added.Results
Tissue parameters are accurately predicted by the neural network mapping, see figures 1 and 2: Root mean square (rms) deviation of T1 and T2 times from true values were 6.6 ms and 3.7 ms respectively. Computation times for calculation of tissue parameters were significantly less for the neural network and could be reduced by 5 orders of magnitude for typical dictionary sizes of 106 entries, see figure 3. Neural networks performed better in terms of robustness against noise: rms deviation of T1 and T2 for the conventional dictionary matching was 13.2 ms and 11.4 ms (for a noise level of 15% of the maximum signal magnitude). For the neural network matching we found mean rms deviations of 10.3 ms and 8.4 ms for the same noise level. An example for a specific tissue (T1= 800 ms, T2= 110 ms) is shown in figure 4.Discussion
In this work we have shown that neural networks provide an efficient and accurate approach to the dictionary matching problem that is both superior in terms of computation time and in robustness to noise. Furthermore this approach can easily be extended to an enlarged input (e.g. very fine dictionary that matches more than three parameters) and to additional output signals (e.g. tissue classification) at almost no additional computational cost.Conclusion
Given the demonstrated advantages of computational efficiency and robustness and additional potential of neural networks in pattern matching, we believe that neural networks will play a major role in reconstruction of magnetic resonance fingerprinting data.Acknowledgements
No acknowledgement found.References
1. Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting. Nature. 2013;495(7440):187-92Figures
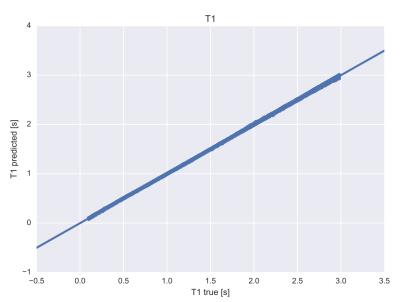
T1
values as predicted by the neural network vs. true values, mean rms deviation
is 6.6ms.
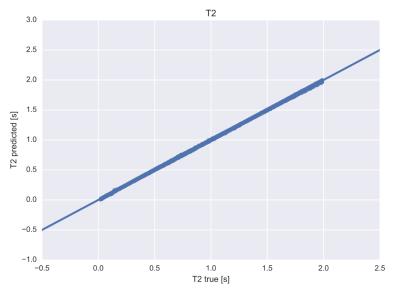
T2 values
as predicted by the neural network vs. true values, rms deviation is 3.7 ms.
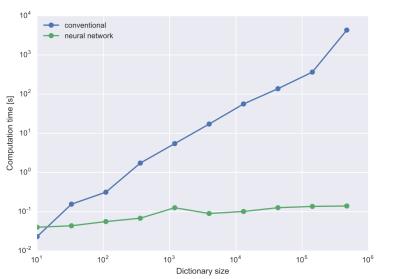
Computation
time for a matching of 500 measurements to a dictionary. The conventional
method is shown in blue and the neural network in green.
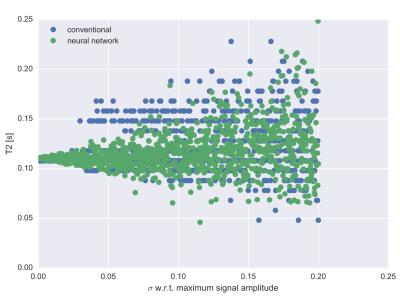
Predicted
T2 values for a tissue with T1=800 ms and T2 = 110 ms vs. added strength of
gaussian noise. Green: neural netword, blue: conventional dictionary matching.