3981
Classification of Head Movements Inside an MRI Scanner using a Single Marker and Neural Networks1JABSOM, University of Hawaii, Honolulu, HI, United States
Synopsis
Detection and classification of head motion may be required for optimal application of prospective motion correction techniques for brain imaging using external tracking systems. Supervised neural networks using various motion metrics were designed to classify head motion inside MR scanner into rigid-body motion and skin motion using single-marker 6-DOF information. The neural networks were trained using volunteer data and then applied to head motion data from 6 clinical in-patients. Neural networks could consistently achieve overall accuracy of 75% or greater.
Purpose
Skin-attached markers used for external motion tracking systems for prospective motion correction (PMC) of MRI are susceptible to non rigid-body motion, e.g. changes in facial expression (“squints”), which can introduce errors in tracking data. We previously proposed the use of multiple markers [1] to detect skin motion, but this may not be feasible in every situation. We previously showed the feasibility of regression models to detect squints, but these algorithms relied heavily on manual tuning [2]. We therefore investigated the use of supervised pattern recognition techniques (neural networks, or NNs) to classify human head motion into skin motion and rigid body motion using single-marker 6-DOF information [3].Methods
Training data for the NNs were obtained from three volunteers that were asked to perform various rigid body motion and non-rigid body motion inside the MRI scanner in a supine position. An MR compatible optical tracking system (Metria Innovation Inc., US, [4]) provided 6-DOF head pose data at 80 samples/second for two separate markers attached to the forehead. The relative rotation between markers was used as the “gold standard” for detecting squints, using a relative rotation of >1°as threshold for “squints”. Motion traces were preprocessed individually and added to the training dataset. NNs were designed using the “pattern-net” algorithm of the Matlab NN toolbox (8.0.1) with scaled conjugate gradient training function and mean square error as performance metric. All networks had the same network parameters and one hidden layer with 18 nodes. The network outputs were classified into two classes, rigid-body motion (RBM) and skin movement (SM). The NN outputs continuous values for RBM and SM, which were converted to binary values using a “winner-takes-all” comparator (larger value was set to 1 and the other 0). The combined training dataset consisted of 25073 samples (RBM = 14,223, SM = 10,800) that were randomly divided into 3 parts for training (60 %), validation (20 %), and testing (20%). Ten networks, each with different training data selected randomly, were designed for each of 4 motion metrics (defined below), for a total for 40 networks. NNs are defined as:
1) NNx – trained using 3 translations and 3 rotations
2) NNsd – trained using standard deviations of 3 translations and 3 rotations
3) NNvel - trained using velocities of 3 translations and 3 rotations
4) NNcc – trained using the 15 correlation coefficients calculated from translations and rotations
The trained NNs were then used to classify the head motion from 6 clinical in-patients undergoing MRI. The study was approved by the local institutional review board. All patients provided verbal and written consent before being enrolled in the study. Motion data were acquired for the entire duration of the clinical brain examination, except when a patient had to be taken out intermittently. No special instructions were given to patients, other than standard instructions not to move. The clinical motion data were pre-processed and passed through all NNs. For each motion metric, the results from the 10 NNs were averaged and converted to binary values.
Results
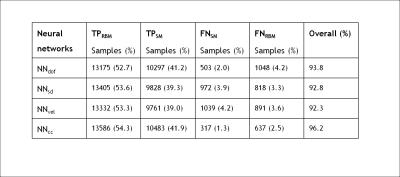
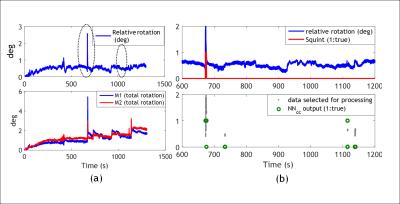
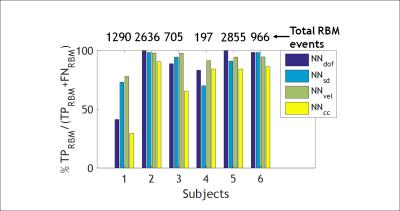
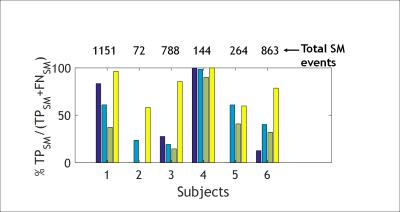
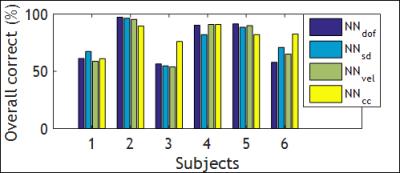
The classification results for the best trained NN for each motion metric are shown in Table I. The best total accuracy is achieved by NNcc. Figure 1 outlines the preprocessing and operation of NNcc on data from clinical patient 4. Figures 2 and 3 show the percentage of data correctly classified as RBM and SM. While NNvel is excellent at detecting rigid-body motion (>90% for 5/6 patients), NNcc is the best of the 4 NN’s at correctly detecting skin motion. The overall accuracy for NNs is defined as ((TPRBM+TPSM)/Total samples). Figure 4 shows that NNcc has an overall accuracy of more than 75% for all but 1 patients.Discussion
As observed, networks tend to bias towards detection of RBM or SM. For our intended application (reacquisition of PMC MR data possibly corrupted with skin motion), missing skin movements (i.e. FNSM) is a more serious error than over-detection of skin movements (FPSM). An occurrence of FPSM will minimally extend the scan (if we reacquire that portion) but the reconstructed images will not be affected. Conversely, an occurrence of FNSM will result in continued motion correction despite skin motion, which may lead to artifacts in the image. Thus,if we consider high TPSM and low FNSM as the ideal characteristic, then NNcc is the best NN for our applications (best performance in squint detection and greater than 75% overall accuracy). Further investigations include analysis of data to determine reasons for mis-classification of data and possible improvements through addition of more training data.Acknowledgements
This work was supported by NIH under grants R01-DA021146 and G12 MD007601.References
1. Singh et.al., “Optical Motion Tracking With Two Markers for Robust Prospective Motion Correction", Magnetic Resonance Materials in Physics, Biology and Medicine, 28(6), pp. 523-534, 2015.
2. Singh et al., "Non Rigid-Body Motion Detection Using Single 6-DOF Data From Skin Based Markers for Brain Imaging", ISMRM 2015.
3. Bishop. C. M, ”Neural Networks for pattern recognition, Oxford University Press.
4. Metria Innovation Inc. (http://www.metriainnovation.com/). Last accessed : Nov 9,2016.
Figures