3980
Fat/Water Classification Using a Supervised Neural Network1GE Global Research, Munich, Germany, 2Global MR Applications & Workflow, GE Healthcare, Madison, WI, United States, 3GE Healthcare, Waukesha, WI, United States, 4GE Global Research, Niskayuna, NY, United States
Synopsis
Fat/Water classification methods relying on image intensity histograms or hydrogen chemical-shift spectra can be subject to failure when assumptions in the algorithm are not met. In this study, we propose a new classification method based entirely on machine learning. Different neural network types were trained and tested on databases covering various anatomies, RF-coil types and image contrasts. A 2D paired classification using a fully connected neural network was capable of reliably classifying fat versus water with an accuracy of 100% on test data sets different from the training data, with a clinically relevant processing time of 0.05 s per case.
Purpose
In Dixon MRI methods, separation of the two chemical species is followed by the step of classification into fat and water images. Algorithms based on evaluating the image intensity of the separated fat and water images, such as histogram methods or methods that exploit the fat chemical shift, have the potential to fail when the intensity characteristics of the images varies significantly due to acquisition protocol, anatomy being imaged, or if a contrast agent is used1,2.
After decades of development, machine learning methods are finally starting to gain widespread traction due to recent improvements in accuracy and speed from new algorithms and hardware, often in combination with cloud computing, making this approach relevant for a number of clinical applications3.
Here we propose to explore the performance of several neural-net architectures applied to fat/water classification.
Methods
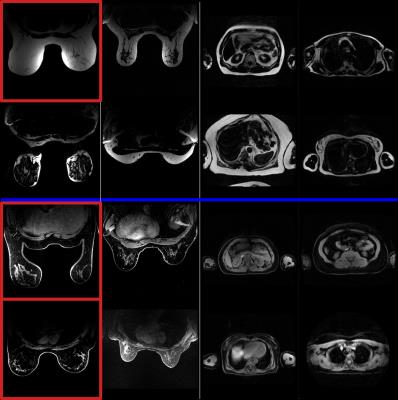
A database of 52 cases was collected on various systems (3T MR750w, MR750, Signa PETMR, 1.5T MR450w, GE Healthcare, WI, USA). All subjects were scanned in the axial orientation with a 3D T1-weighted SPGR with 2-point Dixon sequence. The database comprises two types of anatomy (thorax and breast, with and without contrast) acquired with different RF coils (breast coil array, volumetric body coil), and includes both healthy and pathological cases. Figure 1 illustrates the diversity of the database. All images were manually labelled as Fat or Water by an experienced user (3 cases wrongly classified by the standard classification method were re-labelled).
The database was split into training (80%) and test (20%) data sets. To ensure a common input matrix size, the input volumes were automatically reformatted (by cropping and resampling).
Three main feature variations of the neural network (NN) were tested:
- a fully connected neural network (“Full”) vs. a convolutional neural network (“Conv”)
- independent classifications of each slice (“2D”) vs. volumetric classification (“3D”)
- independent classification of each chemical species as fat or water (“Single”) vs. joint classification of the pair as fat/water or water/fat (“Paired”).
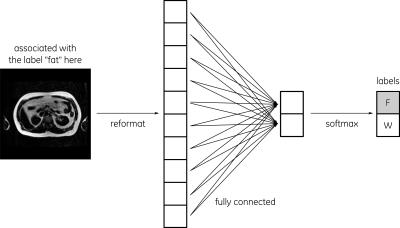
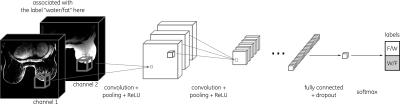
The fully-connected NN was composed of 1 layer with a gradient descent optimizer (Fig. 2) while the convolutional NN was composed of 6 hidden layers (including 2 fully connected) with an Adam optimizer (Fig. 3). All 8 different types of NN were implemented in TensorFlow4. In all cases, cross entropy was used as a cost function.
For each method, the following evaluation criteria were measured: classification accuracy on the test data set, computation time for training, and computation time for classification of one test case.
In order to avoid any bias induced by the database or the random-weight initialization, the 2 steps of training and testing were repeated 3 times in an independent manner after randomly shuffling cases into the training and test data sets. The evaluation criteria were averaged over all repetitions.
Results
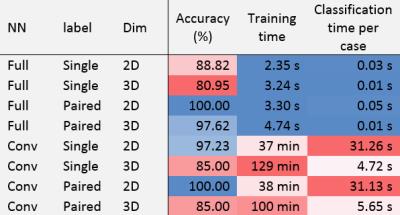
The evaluation criteria are presented in Table 1.
The 2D NN performed systematically better than 3D, convolutional NN better than fully connected NN, and paired classification better than single classification. The following NN reliably resulted in a 100% classification accuracy: {full,paired,2D} with 0.05 s for classification time, and {conv,paired,2D} with 31 s.
Discussion
Overall, convolutional NN were found to be more robust than fully connected NN which is expected since convolution neural networks are independent to the features positions. Also, the number of nodes was higher in the convolutional case.
Paired classification performed better than single, which is expected because more information is provided to the NN to determine the class.
For the same reason, one could have expected 3D to performed better than 2D. The reason why this was not the case may be due to the resampling factor which was higher in the 3D to overcome memory limitation.
Nonetheless, it is interesting to note that the {full,paired,2D} NN was reaching a 100% accuracy with a classification time of 0.05 s. One may expect this accuracy to decrease on a larger and even more diverse database while the accuracy of the {conv,paired,2D} should remain high.
The training time is mainly linked to the number of nodes of the NN, but does not matter in clinical practice since it has to be executed only once for all.
A large number of parameters can be tuned further (initialization, drop off, learning rate, number and size of layers, training batch size, etc.), indicating potential performance improvement opportunities. Such optimization should be performed on an even larger database.
Conclusion
This preliminary study already demonstrates the feasibility of classifying fat/water using a {full,paired,2D} neural network with 100% accuracy and clinically relevant computation time of 0.05 s per case.Acknowledgements
No acknowledgement found.References
1. Ahmad M. et al. A method for automatic identification of water and fat images from a symmetrically sampled dual-echo Dixon technique. Magn Reson Imaging. 2010 Apr;28(3):427-33.
2. Ladefoged C. et al. Impact of incorrect tissue classification in Dixon-based MR-AC: fat-water tissue inversion, EJNMMI Physics 2014 Sept; 1:101
3.Wernick et al. Machine Learning in Medical Imaging. IEEE Signal Process Mag. 2010 Jul; 27(4): 25-38
4. Abadi M. et al. TensorFlow: Large-scale machine learning on heterogeneous systems, 2015. Software available from tensorflow.org.
Figures