3979
Automated reference-free assessment of MR image quality using an active learning approach: Comparison of Support Vector Machine versus Deep Neural Network classification.1Department of Radiology, University of Tuebingen, Tuebingen, Germany, 2Institute of Signal Processing and System Theory, University of Stuttgart
Synopsis
In this study we compare the performance of Support Vector Machine (SVM)-based and Deep Neural Network (DNN)-based active learning for automated assessment of MR image quality. MR images were labeled by radiologists concerning perceived image quality and used as training and test data. DNN and SVM were trained to classify image quality on the training data. An active learning scheme was used for optimization of the training procedure. We found that using acitve learning with either SVM- or DNN- based classification allows for accurate and efficient automated assessment of MR image quality.
Introduction and Purpose
Image quality can be highly variable in MRI due
to numerous influencing technical and patient-dependent factors. The
information content of MR data strongly depends on underlying image quality.
Thus, it is of crucial importance to assess the quality of MR images before
further analysis. This assessment is usually performed visually by radiologists
or MR scientists which makes it time-consuming and subjective. Automated
assessment of MR image quality can provide a solution to this problem that can
be applied in a fast and reproducible manner. First approaches have been
proposed using Support Vector Machine (SVM) – based learning and
classification 1,2,3. In this study, we propose a framework for active learning –based
assessment of MR image quality comparing SVM-based versus Deep Neural Network
(DNN) – based training and classificatio.Methods
The underlying MR data base for training and classification consisted of 2911 2D and 3D data sets from 528 patients and volunteers. Acquisitions of the head/neck, the thorax and the abdomen/pelvis and different sequence types (GRE, SE, FLSH, IR, STIR, EPI) were included. All data sets were acquired on a clinical 3 Tesla PET/MR scanner (Biograph mMR, Siemens Healthineers, Erlangen, Germany). A total of 17386 image features were extracted from each data set (histogram features, textural features, geometrical features, local features). All data sets were labeled by five experienced radiologists on a 5-point Likert scale with respect to overall image quality (1-excellent to 5-non diaganostic). 70 % of the data sets were randomly selected as training data, 30 % as test data. For the purpose of supervised learning and classification, an SVM and a DNN classifier were implemented within this study. The SVM classifier was implemented based on the LIBSVM library 4. 10-fold cross validation was applied for optimization of the parameters C and gamma of the radial basis function kernel. The DNN classifier was implemented using the Keras library for Theano 5. The network consisted of 5 layers (3 hidden layers) with a rectifying linear unit activation function. Weights and bias of each node in the layers are optimized via an adaptive moment estimation with a 10-fold cross validation for learning rate and optimizer settings adaptations.
In order to improve the training step and potentially minimize the number of data sets that need to be labeled manually, we implemented an active learning scheme. Beginning with a small training set of 200 data sets, additional data sets were included into the training setbased on the level of uncertainty of SVM- or DNN- based classification. The level of uncertainty was determined by the class probability differences 6, i.e. small differences yielded a large uncertainty.With the larger training set of size NT the classifiers were updated. Afterwards the next data sets were added to the training set based on the uncertainty until a test set accuracy ACC>90% was reached.
Test set accuracy ACC was defined as the percentage of correctly classified data sets compared to the manually labeled data sets.
Results
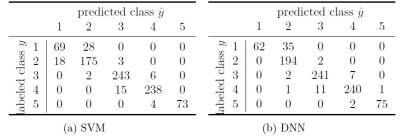
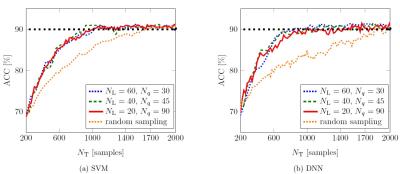
Image labels by radiologists were relatively evenly distributed among the 5 quality classes (Figure 1). Using optimal algorithm parametrization with the full available training data set, overall classification accuracies of 91.3 % and 93 % were achieved using the SVM- and DNN- based classifier respectively (Figure 2). Compared to a random sampling which reflects a normal labeling procedure (no uncertainty-based data set selection), the active learning approach resulted in a markedly steeper learning curve for both, the SVM-based and the DNN-based classifier (Figure 3).Discussion
In this study we propose an active-learning based automated assessment of MR image quality. We evaluated this approach using an SVM-based and a DNN-based classifier and found that high classification accuracy can be obtained with either algorithm while achieving an efficient learning step. In order to further improve the accuracy of this framework, a distinction between different anatomic regions, artifact types and underlying MR sequences will be evaluated in future studies.Conclusion
Automated assessment of overall MR image quality is feasible and yields good classification accuracy using SVM- or DNN- based classification. An active learning approach helps to reduce the amount of needed training labels for both classifiers.Acknowledgements
No acknowledgement found.References
[1] Küstner et al., Proc ISMRM 2015. [2] Küstner et al., Proc ISMRM 2016. [3] Liebgott et al., Proc ICASSP 2016. [4] Chang, ACM T. Intelligent Syst and Tech 2011:2, 1-27. [5] Chollet, Keras on GitHub 2015. [6] Joshi et al.,IEEE Conf. Comp. Vis. Pattern Recogn2009.Figures