3975
In-vivo Segmentation of Carotid Plaque MRI with Deep Convolutional Neural Networks1Institute for Interdisciplinary Information Sciences, Tsinghua University, Beijing, People's Republic of China, 2Center for Biomedical Imaging Research, Tsinghua University, Beijing, People's Republic of China
Synopsis
MRI is gaining popularity for identifying atherosclerosis, a common disease caused by the accumulation of cholesterol in arteries. To identify vulnerable plaque, the components in plaque need to be segmented by radiologist manually, which is both hard and tedious. Previous attempts to solve the problem using probability maps are limited by their accuracy. We leverage the recently developed convolutional neural networks (CNN) to build a model based on 1,000 subjects automatically, achieving significantly better accuracy in almost every metric over traditional methods.
Purpose
It is challenging and tedious for a radiologist to identify and segment plaques in carotid MRI. Previous computer-aided-diagnostics methods, such as MEPPS, heavily depends on heuristics and does not provide enough accuracy. We propose a novel convolutional neural networks (CNN) based method. For hard-to-detect cases such as necrotic/lipid core or hemorrhage, we improved accuracy by over 5x comparing to the state-of-the-art.DataSet
Our data set includes 1,098 subjects (16 locations per subject, and totally 17,568 locations) with institutional review board approval, collected from Center for Biomedical Imaging Research in Tsinghua University. Each case has four MRI sequences T1, T2, TOF and MP Rage. All subjects are viewed by trained radiologists with peer review. Radiologists view and delineate the images with the CASCADE system, 1 identifying tissues including calcifi cation, necrotic/lipid core, hemorrhage, loose matrix as well as fibrous tissue in the data set. Out of the 1,098 subjects, we use 880 for training and 218 for testing.Methods
We formulate the problem of carotid plaque segmentation as a pixel-wise segmentation prediction problem, where we take a location of MRI sequences as input, and automatically outputs a pixel-wise label map, classifying the pixels into di fferent tissues. We use convolutional neural network (CNN) model. Our model is based on residual network. 2 We choose this network because it gives best result on ILSVRC2015 3 detection, recognition, localization and COCO 4 detection and segmentation. We fi ne-tune the residual network with 101 layers pre-trained on ImageNet 5 dataset. Using pre-trained model allows us to obtain the model with reduced number of training images. The model cannot be applied to carotid MRI directly, since the net takes 3-channel RGB images as input and outputs a probability distribution of 1,000 classes. Each input location in our data contains four gray-scale images (T1, T2, TOF and MP Rage), and the output is the pixel-wise segmentation result. In original residual net, the input layer connects with a convolution layer. We make four copies of that convolution layer, and connect each input image with one copy. Then we merge the output of these four convolution layers by summation, and connect the output to the rest of the net. We modify the fully connected layer of residual network to fully convolutional layer and add a deconvolution layer at the end of the network to produce the pixel-wise result. Our modifi ed residual network can take images with arbitrary sizes as input and generate output images of the same size as input.Results
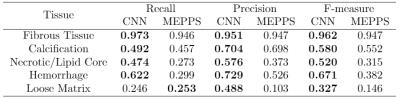
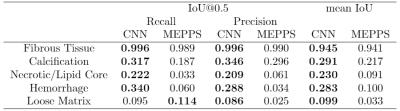
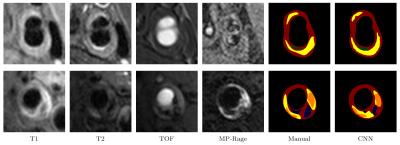
We perform our experiments on a Ubuntu server with a Titan X GPU running Caff e. 6 Training takes about 20 hours while predicting a location takes less than 1 second. We evaluate the predictions with two metrics: 1). We compare our prediction with ground truth (manual segmentation) pixel by pixel and report precision, recall and F-measure for each class. 2). We calculate the intersection over union (IoU=TP/(TP+FP+FN)) for each class in every location, if IoU is no less than 0.5, we consider the prediction to be right. We report precision and recall, defi ned by precision=(correctly predicted locations/positive locations predicted by model) and recall=(correctly predicted locations/positive locations labeled by reviewer). We also report mean IoU, averaged over all positive locations labeled by reviewer or model. We use the segmentation result of MEPPS 7 from CASCADE 1 as our baseline. Table 1 and Table 2 show the pixel-wise segmentation and intersection over union results. CNN performs a little better than MEPPS in fibrous tissue and calcification, and much better in necrotic/lipid core and hemorrhage. Neither methods perform well for loose matrix, due to the lack of positive samples in the training data set. Figure 1 shows some examples of input images and CNN segmentation results.
Conclusion
CNN is a promising tool in automated analysis of MRI images. In particular, it completely trains on example data points, instead of "rules". Even with a moderate sized training set, a carefully tuned CNN outperforms traditional probability methods with lots of heuristics in the problem of carotid plaque segmentation. Also, as we used nearly zero human-knowledge in the process, we can apply the same method to a variety of segmentation tasks in MRI automatic diagnostics.Acknowledgements
No acknowledgement found.References
1. D Xu, WS Kerwin, T Saam, M Ferguson, and C Yuan. Cascade: Computer aided system for cardiovascular disease evaluation. In Proc ISMRM, page 1922, 2004.
2. He K, Zhang X, Ren S, et al. Deep Residual Learning for Image Recognition[J]. Computer Science, 2015.
3. Russakovsky O, Deng J, Su H, et al. ImageNet Large Scale Visual Recognition Challenge[J]. International Journal of Computer Vision, 2015, 115(3):211-252.
4. Lin T Y, Maire M, Belongie S, et al. Microsoft COCO: Common Objects in Context[M]// Computer Vision – ECCV 2014. Springer International Publishing, 2014:740-755.
5. Krizhevsky A, Sutskever I, Hinton G E. ImageNet Classification with Deep Convolutional Neural Networks[J]. Advances in Neural Information Processing Systems, 2012, 25(2):2012.
6. Jia, Yangqing, Shelhamer, et al. Caffe: Convolutional Architecture for Fast Feature Embedding[J]. Eprint Arxiv, 2014:675-678.
7. Fei L, Xu D, Ferguson M S, et al. Automated in vivo segmentation of carotid plaque MRI with Morphology-Enhanced probability maps[J]. Magnetic Resonance in Medicine Official Journal of the Society of Magnetic Resonance in Medicine, 2006, 55(3):659-68.