3951
A Fast Interleaved Bipolar Imaging Method for Fat Quantification1Korea Advanced Institute of Science and Technology (KAIST), Daejeon, Korea, Republic of
Synopsis
The fat quantification using the bipolar multi-echo signals has several benefits such as fast imaging time, SNR, resolution, and robust separation. However, the fat quantification using the bipolar multi-echo signals suffers from the bipolar artifacts due to the imperfect gradient. In this abstract, to overcome these problems, fat quantification is independently performed for each polarity of the readout gradient. Because the acquisition of fully sampled data takes too much time, a new interpolation method for interleaved bipolar multi-gradient-echo acquisition is proposed, which uses the low-rankness of entire data. The experiment results show that the proposed method successfully quantifies the correct fat fraction without bipolar artifacts in a short imaging time.
PURPOSE
Multi-gradient-echo acquisition is widely used for the chemical-shift separation1. Fat quantification is one of the important applications of the chemical-shift separation2. Typically, six echo-times images are required for accurate fat measurement2. Multi-echo acquisition can be performed using either unipolar or bipolar readout gradient. Acquisition with the unipolar readout is preferred because imaging with the bipolar readout includes amplitude and phase error due to eddy current from gradient pulses. However, the bipolar readout acquisition has higher SNR and reduces minimum TR and imaging time. Furthermore, the robust water-fat separation and higher resolution may be achievable by adjusting the echo-spacing time. To exploit the benefits of bipolar readout acquisition, previous works have tried to separate the chemical-shift components using bipolar multi-gradient-echo acquisition by compensating the error between signals from positive and negative gradient3-5. In this abstract, a new bipolar multi-gradient-echo acquisition method is proposed, which independently quantifies fat fraction for each polarity of the readout gradient in a short imaging time.METHODS
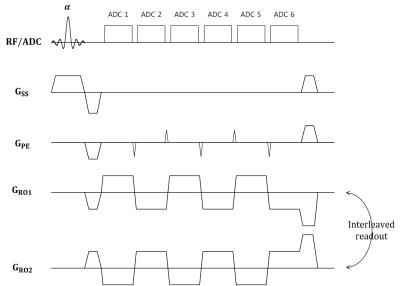
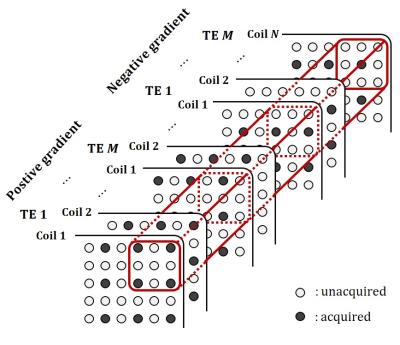
Figure 1 shows that the overall flow diagram of the proposed method. The proposed method acquires data from the interleaved bipolar multi-gradient-echo sequence described in Figure 2. The sequence exploits two bipolar readout gradient trains, which have different initial polarity of readout gradient. The sequence doesn’t acquire the entire multi-echo data from positive and negative readout gradient to save the imaging time. The echo signals from the positive and negative readout gradients and from adjacent echo times are interleaved respectively, each of which is sub-sampled in the phase encoding direction. To estimate entire bipolar multi-echo data from the sub-sampled data for each polarity readout gradient, the new reconstruction method is proposed described in Figure 3. There is similarity between images from the positive and negative gradient and also from different echo times. It implies that there is low-rankness of whole data in k-space domain. Therefore, the low-rankness exploiting method such as GRAPPA6 can be utilized to estimate data from the whole sub-sampled data. In this abstract, the GRAPPA is modified to take whole dimension kernel in k-space. After reconstruction, the chemical-shift separation is performed for the positive and negative readout gradient data, respectively. The chemical-shift separation for each readout polarity prevents generation of bipolar artifacts. The fat quantification is readily performed using the separated water and fat images.
Experiments were conducted on a 3.0 T MRI scanner (Siemens Verio, Germany). The interleaved bipolar multi-gradient-echo imaging was performed using 8-channel knee coil with following parameters: flip angle = 10°; repetition time = 20ms; first echo time = 2.80ms; echo spacing = 1.93ms; receiver bandwidth = 580Hz/pixel; FOV = 180mm×180mm, matrix size = 192×192, slice thickness = 5mm, the number of average = 5, and total imaging time is 19.2 second. The acquired data was sub-sampled retrospectively as R=6, and 24 ACS lines were used so that Reff =3.7 and total scan time is about 5.3 second. The GRAPPA and the proposed reconstruction method were performed with 9×15 kernel.
RESULTS
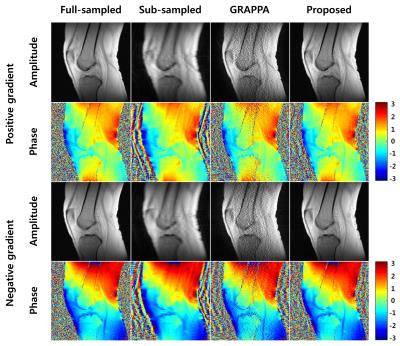
Figure 4 shows the first echo-time images. The reconstructed images from GRAPPA suffer from low SNR and inaccurate phase information due to high acceleration factor. The proposed method reconstructs the magnitude and phase data from both positive and negative readout gradients correctly.
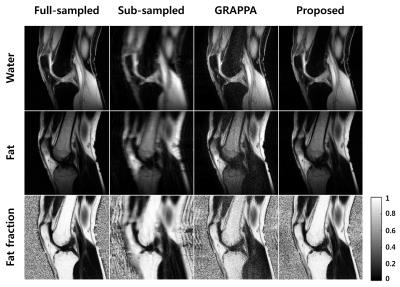
Figure 5 shows the chemical-shift separation and fat quantification results from positive readout gradient in Figure 4. The results from the sub-sampled data suffer from aliasing, low SNR, and incorrect separation. In GRAPPA, there are still low SNR and incorrect separation problems. The result from the proposed method separated water and fat successfully with higher SNR. It implies that the proposed method can reconstruct the correct amplitude and phase of entire echo-time images.
DISCUSSION AND CONCLUSION
The fat quantification using the bipolar multi-echo signals has several benefits such as fast imaging time, SNR, resolution, and robust separation. However, the fat quantification using the bipolar multi-echo signals suffers from the bipolar artifacts due to the imperfect gradient. In this abstract, to overcome these problems, fat quantification is independently performed for each polarity of the readout gradient. Because the acquisition of fully sampled data takes too much time, the interpolation method for interleaved bipolar multi-gradient-echo acquisition is proposed, which uses the low-rankness of entire data. The experiment results show that the proposed method successfully separates water and fat, and acquires the correct fat fraction without bipolar artifacts in a short imaging time. The proposed method can be applied to both 2D and 3D imaging. Furthermore, the proposed method can be extended to other reconstruction methods using low-rankness, such as PRUNO7, SPIRiT8, ESPIRiT9, and SAKE10.Acknowledgements
This research was partly supported by the Brain Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (2014M3C7033999) and Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number : HI14C1135).References
1. Reeder SB, Wen Z, Yu H, Pineda AR, Gold GE, Markl M, Pelc NJ. Multicoil Dixon chemical species separation with an iterative least-squares estimation method. Magn Reson Med 2004; 51: 35–45.
2. Reeder SB, Cruite I, Hamilton G, Sirlin CB. Quantitative assessment of liver fat with magnetic resonance imaging and spectroscopy. J Magn Reson Imaging 2011; 34(4), 729-749.
3. Lu W, Yu H, Shimakawa A, Alley M, Reeder SB, Hargreaves BA. Water-fat separation with bipolar multiecho sequences. Magn Reson Med 2008; 60: 198–209.
4. Yu H, Shimakawa A, McKenzie CA, Lu W, Reeder SB, Hinks RS, Brittain JH. Phase and amplitude correction for multi-echo water–fat separation with bipolar acquisitions. J Magn Reson Imaging 2011; 31: 1264–1271.
5. Soliman AS, Wiens CN, Wade TP, McKenzie CA. Fat quantification using an interleaved bipolar acquisition. Magn Reson Med 2016;75:2000–2008.
6. Griswold MA, Jakob PM,Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 2002; 47: 1202–1210.
7. Zhang J, Liu C, Moseley ME. Parallel reconstruction using null operations. Magn Reson Med 2011;66:1241–1253.
8. Lustig M, Pauly JM. SPIRiT: iterative self-consistent parallel imaging reconstruction from arbitrary k-space. Magn Reson Med 2010;64:457–471.
9. Uecker M, Lai P, Murphy MJ, Virtue P, Elad M, Pauly JM, Vasanawala SS, Lustig M. ESPIRiT-an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA. Magn Reson Med 2014;71:990–1001
10. Shin PJ, Larson PE, Ohliger MA, Elad M, Pauly JM, Vigneron DB, Lustig M. Calibrationless parallel imaging reconstruction based on structured low-rank matrix completion. Magn Reson Med 2014;72:959–970.
Figures