3940
Reconstruction of highly accelerated free-breathing 3D abdominal MRI using Stacked Convolutional Auto-Encoder Network1Academy for Advanced Interdisciplinary Studies, Peking University, Beijing, People's Republic of China, 2Philips Healthcare, Suzhou, People's Republic of China, 3College of Engineering, Peking University, Beijing, People's Republic of China, 4Department of Radiology, Peking University First Hospital, Beijing, People's Republic of China
Synopsis
Free-breathing 3D abdominal imaging is a challenging task for MRI since respiratory motion severely degrades image quality.Our purpose is to develop a novel reconstruction approach for highly accelerated free-breathing 3D abdominal images with stacked convolutional auto-encoders. The whole structure of our proposed method consists of 9 hidden layers except to input and output layer.The proposed method achieves similar quality to the whole sampling reconstruction with non-significant differences for structural similarity index measure (SSIM) (0.99 and 1.00, respectively). Moreover, the average reconstruction time is very short (about 0.25 s/image). Therefore, our method should be employed for a wide range of clinical applications.
Introduction
Free-breathing 3D MRI of the abdomen is a challenging task since respiratory motion can produce image blurring or ghosting artifacts [1]. One of the most commonly used self-navigation technique is 3D golden-radial phase encoding (G-RPE) trajectory sampling with advantages in speed, resolution and free-breathing. But the drawback is that streaking artifacts can be observed clearly from the reconstruction images due to very high undersampling factors.
This work presents a novel method of MRI reconstruction for reducing motion artifact based on Stacked Convolutional Auto-Encoder (SCAE) Network.
Moreover, the long reconstruction time of approaches such as total variation regularized [2] and motion compensation [3] is overcome by our method, which has a promising prospect in clinical use.
Method
The experimental data for training was obtained from ten healthy subjects using a 3D golden radial phase encoding [4] eThrive sequence on a clinical 1.5T MRI system (Ingenia, Philips Healthcare, Best, The Netherlands). The scanner imaging parameters were: slice thickness = 3 mm with over contiguous sampling ; flip angle = 10 degrees; field of view (FOV) = 450 × 450 × 249 mm2; sense factor along z = 1.41; number of readout points in each spoke = 752 with two times oversampling; spatial resolution = 1.00 ×1.00 ×3.00 mm3; repetition time/echo time (TR/TE) = 4.88 ms/2.06 ms. A total of 450 spokes were acquired for each partition, with a total scan time of ~100 s.
The image obtained with 450 spokes is taken as ground-truth image. However, image to be reconstructed by SCAE is consisted of the first 90 spokes, which is full of conspicuous streak artifacts.
Fig. 1 illustrates the whole structure of our proposed method. In addition to the input and output layer, the network is composed of 9 hidden layers: 1) Convolutional layer with 128 5×5 filters; 2) Max-pooling layer of 2×2; 3) Convolutional layer with 64 5×5 filters per map; 4) Max-pooling layer of 2×2; 5) Deconvolutional layer of 64 maps of filter size 5×5; 6) Max-pooling layer of 2×2; 7) Deconvolutional layer of 128 maps of filter size 5×5; 8) Max-pooling layer of 2×2; 9) Deconvolutional layer of one map with filter size 5×5.
Keras[5] was used for implementing this model on a Linux machine (Ubuntu 14.04 LTS; Cuda 7.5). All networks were trained on NVIDIA GPUs (GTX 1080).
The reconstruction accuracy is measured in terms of structural similarity index measure (SSIM). Because the conduct SSIM estimate, already contains luminance, contrast and other remaining errors of images.
Result
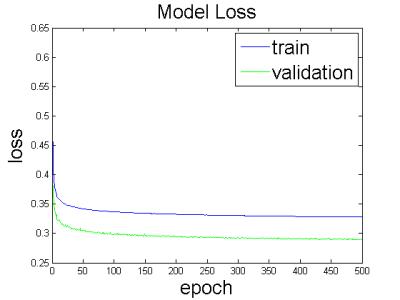
As shown in Figure 2, using a batch size of 1024 and 500 epochs, the loss curves converged nicely. It can be seen that even using 250 epochs, reducing training time in half, we would have got similar results.
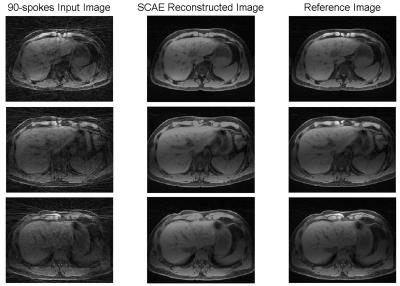
It is clearly seen that strong streaking artifacts existing in Figure 3a are significantly reduced with the proposed approach in Figure 3b.
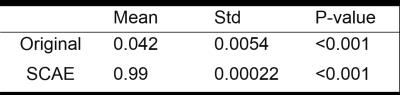
From Table 1, we observe that SCAE yields significant higher SSMI in terms of the input images. In addition, the average reconstruction time is 0.25 seconds per image.
Discussion
Preliminary results demonstrated the feasibility of our approach to correct motion artifacts of highly accelerated free-breathing 3D abdominal MRI. Visual inspection indicates that there are nearly no difference between the SCAE reconstruction and the ground-truth images. The nonlinear relationship between images of two types (streak-contaminated and streak-free) is effectively captured by a large number of trainable mapping and parameters in our network.
Conclusion
In conclusion, this is the first work that proposes to reduce motion artifacts of MRI using stacked convolutional auto-encoder network. Moreover, our method enables reconstruction of one image within seconds and therefore should be employed for a wide range of clinical applications.
Acknowledgements
No acknowledgement found.References
1. Schultz, C.L., et al., The effect of motion on two-dimensional Fourier transformation magnetic resonance images. Radiology, 1984. 152(1): p. 117-21.
2. Cruz, G., et al., Accelerated motion corrected three-dimensional abdominal MRI using total variation regularized SENSE reconstruction. Magnetic resonance in medicine, 2015.
3. Rank, C.M., et al., 4D respiratory motion- compensated image reconstruction of free-breathing radial MR data with very high undersampling. Magnetic resonance in medicine, 2016.
4. Boubertakh, R., et al., Whole- heart imaging using undersampled radial phase encoding (RPE) and iterative sensitivity encoding (SENSE) reconstruction. Magnetic resonance in medicine, 2009. 62(5): p. 1331-1337.
5. Chollet, F., keras. GitHub repository, https://github.com/fchollet/keras, 2015.
Figures