3821
Segment Deep Gray Matter Nucleus from MR Images: An Automatic Computational Tool for Early Diagnosis of Parkinson’s Disease1Department of Radiology and BRIC, The University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 2School of Software, Tsinghua University, Beijing, People's Republic of China, 3Department of Radiology, Capital Medical University, Beijing, People's Republic of China, 4School of Basic Medical Sciences, Fudan University, Shanghai, People's Republic of China, 5Shanghai Key Laboratory of Medical Imaging Computing and Computer-Assisted Intervention, 6Med-X Research Institute, Shanghai Jiao Tong University, Shanghai, People's Republic of China
Synopsis
Accurate and automatic brainstem nuclei segmentation from MR images plays an important role in seeking for imaging-biomarkers of Parkinson’s disease (PD). To address the segmentation challenge from regular MR images, we propose a novel multi-atlas patch based label fusion method where we use hyper-graph technique to handle the low image contrast issue. Our proposed method is successfully applied to a set of MR images from PPMI (Parkinson’s Progression Markers Initiative) dataset, and we have achieved significant improvements in terms of segmentation accuracy compared to the state-of-the-art methods.
Introduction
Based on current neuroscience and clinical findings, there are observable structural alternation at brain stem in the pre-clinical stage of PD. Substantia nigra (SN) and red nucleus (RN) are two small but critical structures which are widely investigated in many PD related neuroscience study and treatment. Hence, accurate and automatic segmentation of these two nuclei can have high impact in various clinical applications, such as deep brain stimulation which aims to debilitate symptoms of Parkinson’s disease (PD), and the investigation of imaging biomarkers, which enables quantitatively measure subtle and complex structural/functional differences related to PD. In regular 1.5T/3.0T T1-weighted MR images, due to the iron deposition during aging, the contrast of SN and RN morphological boundary is very low. Thus, the lack of intensity difference along the structure boundary makes the state-of-the-art multi-atlas patch-based label fusion methods difficult to accurately segment the SN and RN from the MR images.Methods
To address the segmentation challenge from the low-contrast MR images, in this work, we propose a novel multi-atlas segmentation method using deep hyper-graph learning. Hyper-graph learning has shown its superiority in image classification problem by capturing group-wise relationship on a hyper-edge by linking multiple vertices [1]. Here, our proposed method takes the advantage to integrate multi-channel information of spatial voxel-to-voxel relationship and atlas-to-subject relationship. Therefore, our method not only keeps the spatial consistency of the to-be-segmented image but also combines population priors from multiple atlas images. Furthermore, to address the low-contrast image appearance issue, we also compute the high-level context features to measure the complex patch-wise relationship. Here, we turn our method into a deep and self-refining model by using context features from the estimated label probability map. In order to further improve accuracy, we allow more and more predicted subject label vertices with high confidence helping to predict those subject vertices that is hard-to-predict. Such dynamic implementation can further improve the label prediction accuracy.Results
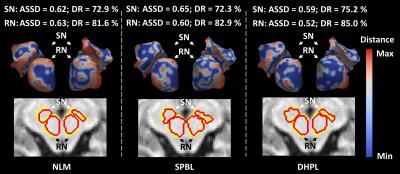
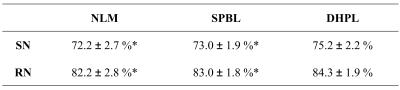
For the experiment, eleven 3.0 Tesla MR images from PPMI dataset are selected. Each subject has the SN and RN manually labeled by two radiologists. To evaluate our method, we applied our proposed deep hyper-graph patch labeling (DHPL) method to automatically segment SN and RN for the 11 subjects, and compared the results with two state-of-the-art label fusion methods: nonlocal mean patch-based label fusion method (NLM) [2], and sparse patch-based label fusion method (SPBL) [3]. To evaluate the performance of our method, we adopted the leave-one-out cross-validation, where each subject was in turn used as a subject image and the remaining 10 subjects were used as atlas images. Table 1 shows the mean and standard deviation of Dice ratios (DR) between ground truth and the estimated segmentations by the three methods. Compared to the SPBL, our method (DHPL) can achieve overall 2.2% and 1.3% improvement in Dice ratio for SN and RN, respectively. Paired t-test further confirmed that the improvement of our method in term of the segmentation accuracy is significant over the two counterpart methods. Fig. 1 further shows the average symmetric surface distance (ASSD) between the manual segmentation and estimated segmentation result by the three methods. Based on the color map shown in the right of Fig. 1, the segmentation result by our DHPL method is much closer to the ground truth than any other two methods.Conclusion
We proposed a novel multi-atlas label fusion method using deep hyper-graph learning to segment the low-contrast brainstem nuclei from the wide available clinical MR images. Our preliminary results show significantly improved segmentation results by using our proposed method compared to the state-of-the-art patch-based label fusion methods, which shows great potential to apply our method in PD early diagnosis.Acknowledgements
This work is supported in part by National Institutes of Health (NIH) grants HD081467, EB006733, EB008374, EB009634, MH100217, AG041721, AG049371, AG042599, CA140413.References
1. Dong, P., Guo, Y., Shen, D., Wu, G.: Multi-atlas and Multi-modal Hippocampus Segmentation for Infant MR Brain Images by Propagating Anatomical Labels on Hypergraph. In: Wu, G., Coupé, P., Zhan, Y., Munsell, B., Rueckert, D. (eds.) First International Workshop, Patch-MI 2015, pp. 188-196. Springer; 2015
2. Coupé, P., Manjón, J.V., Fonov, V., Pruessner, J., Robles, M., Collins, D.L.: Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation. NeuroImage 2001;54, 940-954.
3. Zhang, D., Guo, Q., Wu, G., Shen, D.: Sparse Patch-Based Label Fusion for Multi-Atlas Segmentation. Nice, France: MBIA; 2012.
Figures