3814
Implementation of Pulseq in GPI Lab1Medical Imaging Research Center, Dayananda Sagar Institutions, Bangalore, India, 2Dept. of Radiology – Medical Physics, Medical Center, University of Freiburg, Germany
Synopsis
A major challenge for researchers in designing new pulse sequences is the limitation of needing to know vendor-specific programming environment. The Pulseq tool in Matlab has overcome this issue. This work demonstrates an open source implementation of Pulseq in GPI Lab to further access to Pulseq. Also, this enables a single platform on GPI to integrate the PSD with the rest of the MR research pipeline: simulation, reconstruction, image analysis and visualization. The Pulseq-GPI implementation is demonstrated through a gradient recalled echo. It is capable of designing all sequences/enhancements that Pulseq currently offers.
Introduction
A major challenge for researchers in designing new pulse sequences is the necessity of learning a vendor-specific pulse programming environment. This increases the time from idea to a prototype implementation. Therefore, a Python language-based open-source pulse sequence design tool with a graphical interface is implemented and demonstrated in this work.
Pulseq, an open-source framework developed by Layton et al. (1) currently allows for pulse sequence design in Matlab (MathWorks, Inc) and Jemris (2). The pulse sequences can be exported to a standardized file format (.seq file), which can be executed on nearly arbitrary MR hardware using vendor-specific interpreters. GPI Lab, a Python-based graphical tool for scientific computing developed by Zwart et al. (3) provides tools to visualize, develop and organize scientific algorithms. The tool demonstrated here integrates the simulation, reconstruction and processing, open-source capabilities of GPI Lab with the PSD capabilities of Pulseq.
Methods
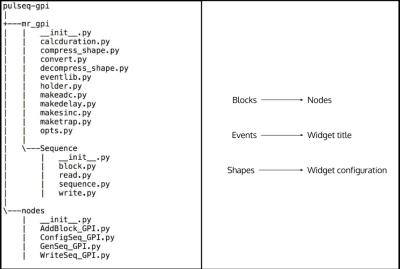
The Pulseq framework includes a custom ‘mr’ toolbox, containing functions, which can be used to design pulse sequences. The toolbox has been ported to GPI Lab, written in Python, called 'mr_gpi’ library. Pulse sequences can be designed in GPI Lab using custom Nodes. These Nodes and the ‘mr_gpi’ library are packaged as ‘pulseq-gpi’ library. The ‘pulseq-gpi’ library code structure is shown in Figure 1(a). The Python implementation closely mirrors the ‘mr’ toolbox’s structure to retain familiarity.
The analogy between Pulseq and GPI Lab is as follows: GPI Nodes are analogous to Blocks, Widget titles are analogous to Events, and Widgets used to configure the Nodes are analogous to Shapes. A mapping of Pulseq’s hierarchical structure to GPI Lab implementation is shown in Figure 1(b).
mr_gpi Library
The ‘mr_gpi’ library requires the Numpy scientific computation library for Python as a dependency, bundled with the GPI Lab integrated development environment.
pulseq-gpi Library
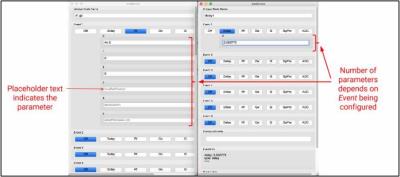
The number of configuration parameters depends on the Event that the user adds to the pulse sequence. For this, a custom GPI Node has been developed, called the ‘AddBlock’ Node. The ‘AddBlock’ Node uses a custom Widget that displays the appropriate number of parameters depending on which Event the user is currently configuring. This is shown in Figure 1(c).
The values computed in each ‘AddBlock’ Node are displayed in the Node, to help the user in configuring the other Events, whose parameters depend on previously added Events.
A custom Python class called ‘Holder’, containing no methods or variables, serves as a replacement to the ‘struct’ data type in MATLAB. Each Event is represented as an instance of the 'Holder’ class, having all the related values encapsulated by that instance.
Gradient Recalled Echo Example
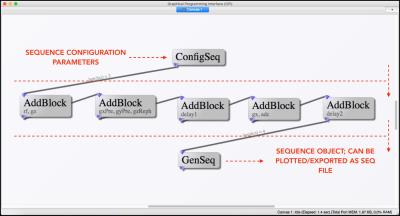
An example network that realizes a Gradient Recalled Echo (GRE) sequence is also included in the library. The GPI Lab network shown has been labeled to provide an easier understanding of how the network works, as shown in Figure 2.
Pulseq File Format
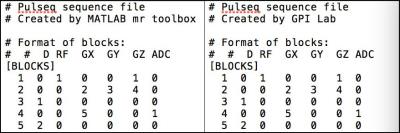
The `pulseq-gpi` library has a custom Node that utilizes ‘mr_gpi’ library’s write functionality to let the user export pulse sequences as a .seq file. This gives the user a single platform to design and export pulse sequences. The .seq files can subsequently be interfaced with MR hardware using vendor-specific interpreter modules. A comparison of the .seq file generated for a GRE sequence by Pulseq’s mr toolbox (implemented using Matlab) and GPI Lab’s network is shown in Figure 3.
Results
Pulseq implemented in GPI Lab reduces the complexity of designing and implementing pulse sequences. Figure 4 shows a comparison of GRE plotted in Matlab and Python. Execution time on MATLAB is 1.8s $$$\pm$$$ 0.1, Python is 2.6s $$$\pm$$$ 0.1 and GPI Lab is 4.7s $$$\pm$$$ 0.1. The delay in execution could be attributed to the multiple function calls between the UI-based Nodes and the back-end code.Discussion and Conclusion
The purpose of this work has been to enable researchers to carry out rapid design and prototyping of pulse sequences, without having to acquaint themselves with vendor-specific code. This tool within the GPI Lab framework enables integration of PSD on a single platform to perform the entire MR research pipeline: simulation, PSD, reconstruction, image analysis and visualization. It does not provide a direct interface (after the PSD stage) to the hardware. However, this is vendor specific and cannot be translated to an open-source tool. Source code can be found at (4).Acknowledgements
Department of Science and Technology (DST), Govt. of India under the program Technology Systems Development (TSD) , No: DST/TSG/NTS/2013/100-G.References
1. Layton, Kelvin J., et al. "Pulseq: A rapid and hardware-independent pulse sequence prototyping framework." Magnetic resonance in medicine (2016).
2. Stöcker, Tony, et al. "High-performance computing MRI simulations." Magnetic resonance in medicine 64.1 (2010): 186-193.
3. Zwart, Nicholas R., and James G. Pipe. "Graphical programming interface: a development environment for MRI methods." Magnetic resonance in medicine 74.5 (2015): 1449-1460.
4. http://www.github.com/sravan953/pulseq-gpi
Figures