3813
An abstraction layer for simpler EPIC pulse programming on GE MR systems in a clinical environment1Neuroradiology, Karolinska University Hospital, Stockholm, Sweden, 2Karolinska Institutet, Stockholm, Sweden
Synopsis
A C-library that simplifies EPIC pulse programming on GE MR systems is presented, containing a large set of new structs and functions for EPIC. Three pulse sequences have been developed (GRE, FSE and EPI) using this library with EPIC. These sequences have most of the functionality expected in a clinical neuro MRI setting, including arbitrary scan planes, full user interface support, online/offline reconstruction, and with common generic sequence plugins/modules such as SpatialSat, FatSat, and Inversion. The KSFoundation library, pulse sequences and sequence modules can be used to quickly develop new pulse sequences with reduced amount of code.
Purpose
Academic researchers modifying the source code of a product pulse sequence can benefit of much of the clinical functionality while repurposing the original sequence. However, this comes at the expense of navigating through a significant amount of code written by others over many years. In contrast, an open source MR pulse programming software, Pulseq, has been presented recently (1), which allows pulse sequences to be designed in Matlab and exported to MR scanners for multiple MR vendors. This is good for testing ideas and for educational purposes, but does not allow complex looping structures or user interactions. A third option is to write ones pulse sequence from scratch in the vendor’s programming environment, typically resulting in a reduced amount of code, but it can be tedious to include fundamental features such FOV offsets, slice ordering, ramp sampling, parallel imaging, SAR safety, sat pulses etc., which may be necessary when a pulse sequence is to be used in a semi-clinical research setting.Methods
Over the last two years we have developed a simplification layer for the EPIC pulse programming environment on GE MR systems. The framework (“KSFoundation”) consists of a C-library containing a large set of object-oriented-like functions and a few common sequence independent files (Fig. 1). This serves to both hide mandatory code (e.g. communicating with the MR hardware) and to simplify the writing of the actual pulse sequence itself. Three pulse sequences have been written from scratch in EPIC using the KSFoundation abstraction layer; ksgre (Gradient Echo), ksfse (Spin Echo and Fast Spin Echo) and ksepi (Echo Planar Imaging), each being about 2-3k lines long, yet having most of the features available in their corresponding product versions. Using the KSFoundation library, the pulse sequence and its pulses are structured as levels of composite structs, making the information to be naturally self contained. New pulse sequence plugins can be added like objects to an existing sequence, making code sharing between programmers in different projects straightforward. RF scaling, SAR, hardware heating limitations as well as TR timing are automatically invoked by dry-running the collection of sequence modules each time the user interface (UI) is changed. Using the handles to the sequence objects, plotting (PNG, PDF) and movie making (animated GIFs) of the sequence can be performed automatically, which is useful for debugging as well as for use in presentations. Furthermore, sequence modules written using the KSFoundation functions can be combined with an existing product pulse sequence. The sequences developed have been scanned on healthy volunteers together with their corresponding product sequences using identical scan parameters, after informed consent and IRB approval.Results
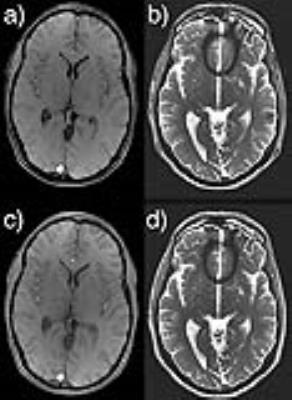
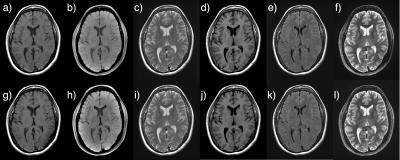
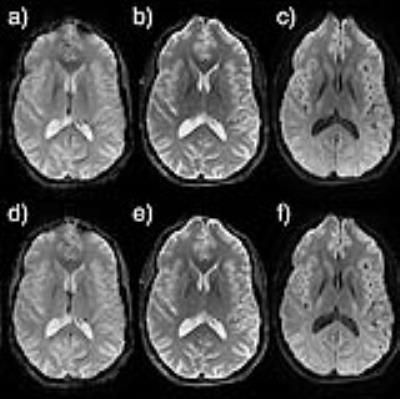
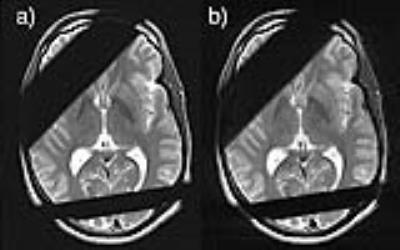
Fig. 1 illustrates the KSFoundation simplification layer for EPIC, noting that the EPIC compilation process is the same after having included the static library. In Fig. 2, two gradient echo sequence variants are shown using a,c) RF spoiling, b,d) bSSFP, where a-b) were made with ksgre and c-d) the product versions. In Fig. 3, six MR contrast are shown for the ksfse sequence including a,g) T1-w, b,h) T1-w with FatSat c,i) T2-w, d,j) T1-FLAIR, e,k) T2-FLAIR and f,l) T2-STIR, where a-f) were made with ksfse and g-l) the product versions. Echo planar images are shown in Fig. 4 in a similar layout with a,e) Gradient echo EPI, b,f) Spin-Echo EPI, c,g) Diffusion-weighted EPI, and d,h) T2-FLAIR EPI. Finally, as an illustration of the spatial saturation plugin made using KSFoundation, Fig. 5 shows a T2-w image prescribed with two spatial saturation bands, with a) the product sequence, b) ksfse.Discussion
We have developed a simplification layer for EPIC, which enables an object oriented-like structure while being compatible with the standard EPIC code. We have made sequences with similar clinical functionality (for neuro MRI) as the corresponding product sequences, using a fraction of the amount of code. The KSFoundation library includes an extensive documentation where one can also view the source code for all library functions and is available on GitLab (2) for free to anyone with a research agreement with GE Healthcare.Acknowledgements
No acknowledgement found.References
1. Layton KJ, Kroboth S, Jia F, Littin S, Yu H, Leupold J, Nielsen J-F, Stöcker T, Zaitsev M. Pulseq: A rapid and hardware-independent pulse sequence prototyping framework. Magn. Reson. Med. 2016. DOI: 10.1002/mrm.26235.
2. Stefan Skare, Enrico Avventi, Ola Norbeck, Henric Rydén. KSFoundation Git repository. https://gitlab.com/neuromr_karolinska/psd_ksfoundation
Figures