3812
Implementation of a parallel processing pipeline of multi-channel phase data1Imaging Research Laboratories, Robarts Research Institute, London, ON, Canada, 2Dept. of Medical Biophysics, Schulich School of Medicine & Dentistry, Western University, London, ON, Canada
Synopsis
Evidence exists to show that phase unwrapping performed before channel combination results in fewer artifacts (singularities, open ended fringe lines) than when phase unwrapping occurs after channel combination. We have implemented a fast and efficient pipeline designed to enable processing of multi-channel phase data. Specifically, non-iterative phase unwrapping and channel combination are employed within the pipeline that links the MR scanner to a DICOM server, which displays the final combined images, while preserving all metadata.
Introduction
Evidence exists to show that phase unwrapping performed before channel combination results in fewer artifacts (singularities, open ended fringe lines) than when phase unwrapping occurs after channel combination.1,2 However, performing pre-channel processing increases the disk storage, memory, and computation requirements by orders of magnitude. For example, the uncombined data of an 80-slice 10-echo 3D gradient echo (GRE) acquisition with a 64 channel array would require 25 GB of storage space and the associated increase in memory and processing time. Routine research and clinical practice require reconstructed data to be available for visualization on a time scale similar to the acquisition time. To this end we have implemented a fast and efficient pipeline designed to enable processing of multi-channel phase data. Specifically, non-iterative phase unwrapping3 and channel combination1 are employed within the pipeline that links the MR scanner to a DICOM server, which displays the final combined images, while preserving all metadata.Methods
Image acquisition
Ten-echo 3D GRE images of the head and neck of ten patients and volunteers were acquired on a 3T scanner with a 64 channel head and spine array coil; the number of channels used for each scan was automatically determined by the scanner software and varied between 14 and 64 depending on the anatomy scanned. The number of slices varied between 36 and 96. The institution’s research Ethics Board approved this research.
The Pipeline
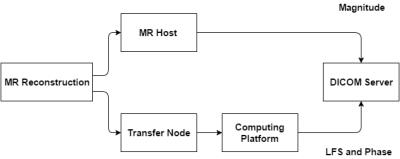
Figure 1 is a block diagram of the established pipeline. The first step is an algorithm implemented in the scanner’s reconstruction environment that results in complex channel images being sent, via a dedicated 1 Gb link, to a transfer node which saves the data in ISMRMRD4 format on an off-line computing platform; timestamps are appended to each filename to ensure uniqueness. Once the transfer is complete, a 'diagnostic' program ensures the data and metadata are in agreement.
A configuration file is generated from a template and the files are passed to a Gadgetron5 instance. The process consists of loading the data from the ISMRMRD file and running it through the pipeline one multi-channeled slice at a time. Phase is unwrapped using the PUROR method,3 high-pass filtered then all echoes of a single slice are collected to perform the channel combination by calculating the inter-echo variance on a pixel-by-pixel basis and using it as the weighting factor.1 At this stage the channel phase data are further processed to calculate local frequency shifts.
Finally, the pipeline saves the combined channel phase and local frequency shift images in DICOM format, preserving the relevant metadata, and transfers them to a DICOM server for viewing and analysis. Alternatively the files can be saved locally as a new ISMRMRD file for easy implementation into additional analysis and processing pipelines.6
Implementation
The pipeline was implemented in C++ and parallelized using OpenMP (https://gcc.gnu.org/wiki/openmp) on a 4-core Intel i7-4770 computer (3.40 GHz) with hyperthreading and 3.4 GB RAM. To ensure cross-platform compatibility the Gadgetron network transfer protocol was modified to transform data to network byte order for all communication.
Evaluation
The accuracy and robustness of the ISMRMRD/Gadgetron processing was compared to a verified implementation of the same algorithms in MATLAB. A range of data sets with different number of echoes and slices were evaluated. The processing times were also measured.
Results
Figure 2 is a comparison of a single local frequency shift map calculated via the pipeline and by MATLAB. The images are nearly identical, with an average absolute difference of <1 Hz within the brain. For a 15.6 GB data set (58 channels, 96 slices, 10 echoes) the pipeline processing time was 230 s, compared to 3088 s using MATLAB. A similar time reduction was achieved with a smaller data set (3.1 GB, 14 channels, 80 slices – 52 s with the pipeline and 506 s with MATLAB).Discussion
The implemented pipeline successfully calculates quantitative maps via channel-by-channel processing of phase data in times that are comparable to the scan time. Quantitatively, agreement with the “gold standard” MATLAB results was excellent but the calculations were achieved in a fraction of the processing time. More importantly, the final quantitative images were ready for routine viewing within an hour of scan completion. Processing time can be reduced further by using more processors or more efficient programming. For the data sets tested, transfer between the reconstruction engine and the transfer node were short and did not interfere with scan acquisition. However for larger data sets, acquired more rapidly this may become a problem, which can be addressed via the introduction of an ultra-high speed connection.Acknowledgements
Partial funding for this work was provided by the Ontario Research Fund. M.D. is a Career Investigator of the Heart and Stroke Foundation of Ontario. This work was made possible by the facilities of the Shared Hierarchical Academic Research Computing Network (SHARCNET:www.sharcnet.ca) and Compute/Calcul Canada. The authors thank H. Hosseini for help with figure preparation.
The processing pipeline code is available for research use online at https://github.com/TWhelan3/PUROR_IEV
References
1. Liu J, Rudko DA, Gati JS, Menon RS, Drangova M. Inter-echo variance as a weighting factor for multi-channel combination in multi-echo acquisition for local frequency shift mapping. Magn Reson Med. 73(4):1654-1661. 2015
2. Hosseini Z, Liu J, Solovey I, Menon RS, Drangova M. Susceptibility-weighted imaging using inter-echo-variance channel combination for improved contrast at 7 tesla. J Magn Reson Imaging. 2016 (epub)
3. Liu J, Drangova M. Intervention-based multidimensional phase unwrapping using recursive orthogonal referring. Magn Reson Med. 68(4):1303-1316. 2012
4. Inati SJ, Naegele JD, Zwart NR, et al. ISMRM Raw data format: A proposed standard for MRI raw datasets. Magn Reson Med. 2016
5. Hansen MS, Sorensen TS. Gadgetron: an open source framework for medical image reconstruction. Magn Reson Med. 69(6):1768-1776. 2013
6. Liu J, Drangova M. Method for B0 off-resonance mapping by non-iterative correction of phase-errors (B0-NICE). Magn Reson Med. 74(4):1177-1188. 2015