3690
3D Hyperpolarized C-13 EPI with Calibrationless Parallel Imaging1Radiology & Biomedical Imaging, UCSF, San Francisco, CA, United States
Synopsis
With the translation of hyperpolarized 13C to the clinic, imaging approaches will require large volumetric FOVs to support clinical applications. Parallel imaging techniques will be crucial to increasing volumetric scan coverage while minimizing RF requirements and temporal resolution. Calibrationless parallel imaging approaches are well-suited for this application because they eliminate the need to acquire coil profile maps or auto-calibration data. In this work, we explored the application of calibrationless parallel imaging (SAKE) and corresponding sampling strategies to accelerate and undersample hyperpolarized 13C data using 3D blipped EPI acquisitions and multichannel receive coils.
Introduction
Dissolution dynamic nuclear polarization provides a 10,000+ fold enhancement to nuclear spin polarization. This huge increase in polarization has enabled non-invasive, real-time metabolic imaging in both pre-clinical (1) and clinical (2) applications. However, the transient nature of the hyperpolarization leads to tradeoffs between spatial resolution, temporal resolution, and SNR. Parallel imaging can be employed to improve coverage while reducing the total scan time and RF requirements. However, acquiring coil profile maps directly from 13C is an inefficient use of the hyperpolarization, and acquiring sufficient autocalibration data becomes more onerous and limits the total acceleration for the relatively small matrix sizes used for 13C acquisitions. Instead, calibrationless parallel imaging techniques such as SAKE (3, 4) are appealing because they eliminate these downsides by exploiting the inherent low-rankness of multichannel data. In this work, we explored the application of SAKE to accelerate and undersample hyperpolarized 13C data using a 3D EPI acquisition and multichannel reception.
Methods
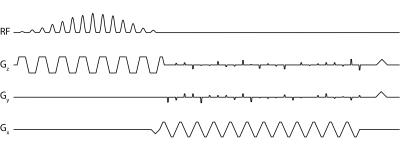
All data were acquired on a GE 3T scanner (MR750, Waukesha WI, USA) with clinical performance gradients (5 G/cm gradient strength, 20 G/cm/ms slew-rate). A 3D echo-planar imaging sequence with blip gradients on both Y and Z (Fig. 1) was used for data acquisition, and an 8-channel coil comprised of two 4 element paddles (5) was used for reception.
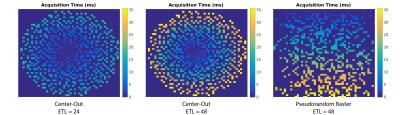
Simulations: Digital simulations were performed in Matlab R2015b. Two different blip schedules were investigated: a center-out acquisition and a pseudorandom raster (6), with regular blips on one axis and pseudorandom blips on the other (Fig. 2). The encoding scheme was designed for a 48 x 48 x 48 matrix size, with ETL = 24 or 48. T2* decay and chemical shift were included in the signal model to quantify the effect of signal decay and off-resonance on the reconstruction.
Phantom/In Vivo Experiments: For 13C phantom and in-vivo acquisitions, data were acquired with a ramp-sampled, symmetric EPI readout with blips on both the Y and Z gradients. Data were acquired on a Cartesian grid, eliminating the need for NUFFT or gridding. The Orchestra toolbox (GE Healthcare) was used for ramp-sampling and reference scan correction, and data were subsequently reconstructed using SAKE on a slice-by-slice basis. Scan parameters for the phantom data are: FOV = 33.6cm3, matrix = 48 x 48 x 48, TR = 62.5ms, NEX = 120, total scan time = 6 minutes. For the in vivo study, t-butanol was polarized in a Hypersense for 60 minutes and injected via tail vein over 15s. Data acquisition started at the end of injection. Scan parameters for the in vivo dataset are identical except for FOV = 32cm3, TR = 78ms, NEX = 1.
Results & Discussion
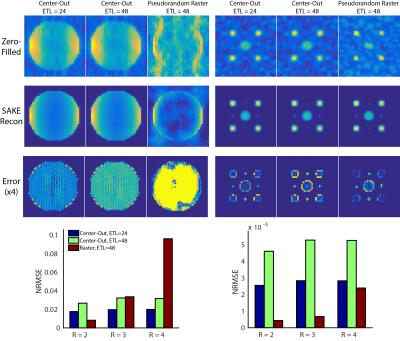
Phantom simulation results for the two blip schedules with different acceleration factors are shown in Fig 3. While the pseudorandom raster was more robust to T2* decay, its overall acceleration was limited, breaking down for R = 4 when the object comprises the majority of the FOV. Conversely, the center-out approach allowed for more flexible sampling, including corner cutting and variable-density sampling. Most of the error for the center-out approaches appeared at the object boundary. Similar to a radial acquisition, the PSF for the center-out blip pattern (data not shown) has an increased peak height and broader width, resulting in higher SNR at the expense of blurring.
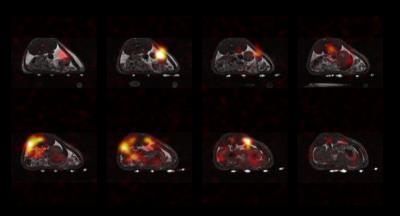
The digital simulations were corroborated by the 13C ethylene glycol phantom experiments in Fig. 4. The center-out scheme with ETL = 24 provided a two-fold reduction in scan time and RF deposition while yielding a beneficial tradeoff between blurring and SNR. For longer T2* relaxation times the ETL can be increased to further improve the temporal resolution. This pattern also reduces the blip step size between echoes, minimizing potential eddy current artifacts. The 3D hyperpolarized t-butanol acquisition with the center-out, ETL = 24 scheme (Fig. 5) is free of Nyquist ghosting and undersampling artifacts, validating the feasibility of 3D EPI and SAKE reconstruction in vivo. Depending on object sparsity and coil configuration further acceleration is likely possible, and will be explored in future work.
Conclusion
Parallel imaging approaches are crucial to increasing the scan coverage for human hyperpolarized 13C MR studies while minimizing the temporal resolution and RF requirements. The 3D EPI sequence developed in this project provided a flexible acquisition that allows for arbitrary 2D undersampling patterns in the two blip (phase-encode) dimensions. The center-out acquisition scheme reduced the TE and improved SNR while providing fourfold undersampling with an eight channel coil. However, T2* must be considered when determining the ETL and total readout time to minimize blurring.Acknowledgements
This work was supported by NIH grants R01EB017449, R01CA183071 and P41EB013598.References
1.) Albers, M.J., et al., Hyperpolarized 13C Lactate, Pyruvate, and Alanine: Noninvasive Biomarkers for Prostate Cancer Detection and Grading. Cancer Res., 2008. 68(20): p. 8607-8615.
2.) Nelson, S.J., et al., Metabolic Imaging of Patients with Prostate Cancer Using Hyperpolarized [1-13C]Pyruvate. Science Translational Medicine, 2013. 5(198): p. 198ra108.
3.) Shin, P.J., et al., Calibrationless parallel imaging reconstruction based on structured low-rank matrix completion. Magnetic Resonance in Medicine, 2014. 72(4): p. 959-970.
4.) Feng, Y., et al., Development and testing of hyperpolarized 13C MR calibrationless parallel imaging. Journal of Magnetic Resonance, 2016. 262: p. 1-7.
5.) Tropp, J., et al., Multi-channel metabolic imaging, with SENSE reconstruction, of hyperpolarized [1-13C] pyruvate in a live rat at 3.0 tesla on a clinical MR scanner. Journal of Magnetic Resonance, 2011. 208(1): p. 171-177.
6.) Geraghty, B.J., et al., Accelerated 3D echo-planar imaging with compressed sensing for time-resolved hyperpolarized 13C studies. Magnetic Resonance in Medicine, 2016.
Figures