3537
The Neonatal DTI fiber atlas for studies of brain development at birth1Center for Investigating Healthy Minds, University of Wisconsin-Madison, Madison, WI, United States, 2Psychology, University of Wisconsin-Madison, Madison, WI, United States, 3Psychiatry, University of North Carolina at Chapel Hill, 4Kitware Inc., 5Waisman Center, University of Wisconsin-Madison, 6Tandon School of Engineering, Department of Computer Science & Engineering, NYU, 7Department of Computer Science, University of North Carolina at Chapel Hill
Synopsis
Given the increasing popularity and wealth of DWI data in the field of neuroimaging, there is a critical need for the development of publically available resources that enable widespread application of a set of template fibers for atlas based along-tract analysis supporting an adequate and reliable analysis of DTI in newborns in both practice and in clinical research settings. To address this gap, we developed a Neonate DTI atlas that represents a typically developing human brain during the first few weeks of life. To the best of our knowledge, we are the first to develop a population atlas with this magnitude of quality and sample size, as well as with a comprehensive set of template fibers for semi-automatic tract based analysis. The DTI atlas and the tracts will be made available through NITRC.
Contributions
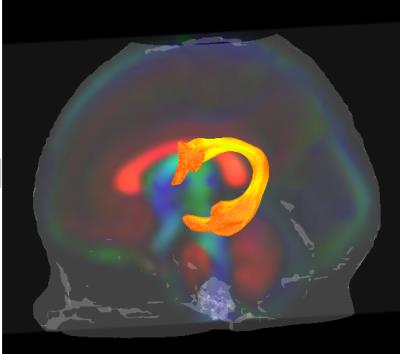
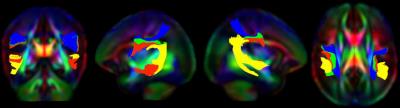
We have created the first atlas to represent a standardized template of the typically developing human brain within the first few weeks of life. Further, we provide the template white matter tract segments for semi-automatic atlas fiber tract based DTI analysis. To the best of our knowledge, we are the first to develop a neonatal healthy population atlas with this magnitude of quality and sample size, as well as with publically available resources that enable for the widespread application of a semi-automatic tract-based analysis. The DTI template, consisting of a diffusion tensor atlas as well as 47 white matter tract segments, may be obtained by visiting NITRC page.Introduction
DTI is a tool that is useful for non-invasively tracing white matter fiber pathways in the human and animal brain. The use of an atlas for a common reference space with a standardized reference for anatomical guidance is increasingly becoming essential in the field of imaging research. Most such atlases to date have been developed for adult human populations and few for non-human primates3,4. In previous DTI studies in newborns2, many rely on the existing atlases of adults or children that are available publically or construct a study specific template from a small sample. There has been minimal, but valuable effort devoted to creating MRI atlases for whole-brain analysis from conventional T1- and T2- weighted images and scalar maps derived from tensor data (1). However, the only public atlas for neonates supports region-based analysis which mixes multiple white matter tract populations within a single region of interest. Additionally, existing atlases are created from small samples and therefore limit registration accuracy. (2) DT Tractography (DTT) has been an effectively valuable tool for probing white matter trajectories as it provides investigations of structural brain correlates with behavior and function(5). Despite the wealth of applications to the investigation of neuronal development and functional maturation, the fiber-tract based analysis approach is not widely applied in the study of neonatal DTI data. The limited availability of appropriate existing brain atlases and the lack of reproducible and reliable tract tracing protocols to date creates the critical need of a human neonatal DTI template with expert-validated fiber tracts that would enable scientists to perform standardized statistical analysisMethods
The DTI atlas was constructed from 144 newborns drawn from a larger, uniquely extensive dataset of nearly 1000 subjects 3 prospective longitudinal neuroimaging studies. The 144 newborns (gestational age at birth between 30 and 42 weeks (37.11 ± 2.6)) were scanned using 3T Siemens model scanner. The atlas included equally representative samples from all three imaging protocols available (Table 1). 85% of the atlas was healthy, while the other 15% had the following conditions (Table 2).
The optimal framework for atlas construction and white matter tractography inherently depends on 1. comprehensive diffusion imaging quality control, 2. accurate atlas building procedures and 3. time intensive interactive tractography in the atlas using hypothesis-driven criterion for white matter tract dissection.
Pre-processing: See Figure 1. This comprehensive assessment process is approximately 30 min. for each subject, is performed to ensure the data was free from most prevalent issues in infant DWI sequences: artifacts both slice-wise and gradient-wise, related to both intensity as well as introduced by subject motion and scanner table vibration.
Atlas Construction: DTI-Reg performs a deformable registration of the DTI datasets using a pairwise registration method. A sequence of processing steps, specifically affine followed by deformable registration, provides significantly better registration results between two DTI datasets. Since the method generates an atlas which is significantly more representative of the healthy newborn population than the traditional study specific template, better white matter fiber tractography results can be expected from tracts generated in the atlas space.
White matter fasciculi were derived from an atlas-based tractography approach using 3D Slicer. Major fiber bundles were separated into “tract segments” based on trajectories to predefined cortical surface targets that correspond to putative functionally related regions. In all, a total of 47 “tract segments” were obtained from the neonatal atlas for further analysis.
Discussion
As a result of our work, we have provided the research community with the resources and framework to reliably and efficiently apply quantitative tract based analysis in the newborn human brain. This atlas based set of template fibers customized to the neonatal population will provide a common analytic framework for quantification of DTI measurement across studies.
Acknowledgements
We would like to acknowledge the following funding sources MH086633, P50 MH064065, MH070890, HD053000, Roadmap Grant U54 EB005149-01, P50 MH078105-01A2S1, UNC Intellectual and Developmental Disabilities Research Center P30 HD03110, MH091645. NIH grants R01-EB02883, UL1RR025011, P30-HD003352.References
Oishi, K. et al. Multi-contrast human neonatal brain atlas: Application to normal neonate development analysis. Neuroimage 56, 8–20 (2011).
Zhang, Y. et al. A Bayesian approach to the creation of a study-customized neonatal brain atlas. Neuroimage 101, 256–67 (2014).
Verde, A. R. et al. UNC-Utah NA-MIC DTI framework: Atlas Based Fiber Tract Analysis with Application to a Study of Nicotine Smoking Addiction. Proc. SPIE 8669, (2013).
Verde, A. R. et al. UNC-Utah NA-MIC DTI framework: atlas based fiber tract analysis with application to a study of nicotine smoking addiction. in SPIE Medical Imaging 86692D--86692D (2013). 1. Farzinfar, M. et al. Diffusion imaging quality control via entropy of principal direction distribution. Neuroimage 82, 1–12 (2013).
Wang, Y. et al. DTI registration in atlas based fiber analysis of infantile Krabbe disease. Neuroimage 55, 1577–1586 (2011).
Gilmore, J. H. et al. Prenatal and neonatal brain structure and white matter maturation in children at high risk for schizophrenia. Am. J. Psychiatry 167, 1083–1091 (2010).
Goodlett, C. B., Fletcher, P. T., Gilmore, J. H. & Gerig, G. Group analysis of DTI fiber tract statistics with application to neurodevelopment. Neuroimage 45, S133--S142 (2009).
Goodlett, C., Corouge, I., Jomier, M., Gerig, G. & others. A quantitative DTI fiber tract analysis suite. Insight J. (2005).
Oguz, I. et al. DTIPrep: quality control of diffusion-weighted images. Front. Neuroinform. 8, 4 (2014).
REFERENCED OPEN SOURCE SOFTWARE 3D Slicer: http://www.slicer.org DTIPrep: http://www.nitrc.org/projects/dtiprep/ itk-SNAP: http://www.itksnap.org/pmwiki/pmwiki.php DTIAtlasBuilder: http://www.nitrc.org/projects/dtiatlasbuilder/ MriWatcher: http://www.nitrc.org/projects/mriwatcher/ DTI-Reg: http://www.nitrc.org/projects/dtireg/ ANTS: http://www.picsl.upenn.edu/ANTS/ FiberViewerLight: http://www.nitrc.org/projects/fvlight/ DTIAtlasFiberAnalyzer: http://www.nitrc.org/projects/dti_tract_stat FADTTS: http://www.nitrc.org/projects/fadtts/
Figures