3533
A unified framework for upsampling and denoising of diffusion MRI data1Image Sciences Institute, University Medical Center Utrecht, Utrecht, Netherlands
Synopsis
Diffusion MRI suffers from relatively long scan times and low signal to noise ratio (SNR), which limits the acquired spatial resolution. In this work, we propose a unified framework for denoising and upsampling diffusion datasets based on a sparse representation of the diffusion signal. Our proposed method shows less blurring and increased anatomical details in the pons region when compared to denoising and subsequent spline interpolation. At the junction of the corpus callosum, the corticospinal tract and the cingulum, finer structures are also preserved as evidenced by a high resolution invivo acquisition.
Introduction
Diffusion MRI (dMRI) suffers from relatively long scan times and low signal to noise ratio (SNR), which limits the acquired spatial resolution. Many techniques have emerged to increase image quality and achieve higher spatial resolutions such as specialized acquisition schemes1, 2, 3, image processing techniques to increase spatial resolution4,5,6 and denoising techniques to increase the SNR7,8,9. While these methods typically aim to either increase the spatial resolution or improve the SNR, it would be beneficial to combine both, enhancing their effectiveness. In this work, we propose such a unified framework for denoising and upsampling dMRI data based on a sparse representation of the diffusion signal.Theory
Sparse linear models have been used as a powerful method to represent the MRI signal by only using a few elements. Such a representation has been used for structural MRI upsampling10 or dMRI denoising8. The key idea is to decompose locally $$$n$$$ dMRI signals $$$X_i$$$ as a linear combination of a few basis elements11 through equation 1. By exploiting the redundancy of the signal, a sparse representation $$$\hat{X}_i=\textbf{D}\alpha_i$$$ can be found, with $$$X_i$$$ the MRI signal in a neighborhood $$$i$$$, $$$\textbf{D}$$$ the dictionary, $$$\alpha_i$$$ the sparse coefficients, $$$\lambda$$$ a tradeoff parameter between data fitting and sparsity and $$$\hat{X}_i$$$ the reconstructed signal.
$$\text{Equation 1}\enspace\min_{\mathbf{D},\alpha}\frac{1}{n}\sum_{i=1}^n\left(\frac{1}{2}\|X_i-\mathbf{D}\alpha_i\|_2^2+\lambda\|\alpha_i\|_1\right)\enspace\text{s.t.}\enspace\|\mathbf{D}\|_2^2=1$$
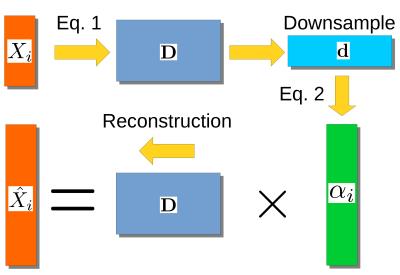
In our unified framework, we propose to extract a lower resolution representation $$$\textbf{d}$$$ by averaging the columns of $$$\textbf{D}$$$ to form smaller patches, thus giving a one-to-one mapping between low and high resolutions patches within a multiscale approach12. We then solve equation 2 using $$$\textbf{d}$$$ with $$$\lambda_i=\sigma^2_i(m+3\sqrt{2})$$$ and $$$m$$$ the number of elements in a patch. This would give a denoised representation8 for $$$\hat{X}_i=\textbf{d}\alpha_i$$$, but by instead using the direct relationship between $$$\textbf{d}$$$ and $$$\textbf{D}$$$, we reconstruct a high resolution, denoised representation $$$\hat{X}_i=\textbf{D}\alpha_i$$$ as shown schematically in figure 1.
$$\text{Equation 2}\enspace\min_{\alpha_i}\|\alpha_i\|_1\enspace\text{s.t.}\enspace\frac{1}{2}\|X_i-\mathbf{d}\alpha_i\|_2^2\le\lambda_i$$
Methods
Two datasets of the same subject were obtained on the same 3T Philips scanner; a 1.8 mm dataset with $$$64\thinspace\text{b}=1000\thinspace\text{s/mm}^2$$$ volumes, $$$\text{TR/TE}=18.9\thinspace\text{s}\thinspace/\thinspace104\thinspace\text{ms}$$$ and a 1.2 mm dataset with $$$40\thinspace\text{b}=1000\thinspace\text{s/mm}^2$$$ volumes, $$$\text{TR/TE}=11.1\thinspace\text{s}\thinspace/\thinspace63\thinspace\text{ms}$$$, both with one $$$\text{b}=0\thinspace\text{s/mm}^2$$$ volume. The 1.2 mm dataset serves as a gold standard for spatial resolution attainable with enough signal within a reasonable timeframe on a standard scanner.
Using the 1.8 mm dataset, we first estimated the variance of the Rician noise with PIESNO13 and corrected for Rician noise bias14. We extracted spatial patches of size (6,6,6) with 5 angular q-space neighbors to construct $$$\textbf{D}$$$ using equation 1 and then downsampled $$$\textbf{D}$$$ by a factor of 2 in each dimension, thus creating $$$\textbf{d}$$$ of size (3,3,3). The coefficients $$$\alpha_i$$$ were computed with equation 2 and $$$\textbf{d}$$$, thus obtaining an upsampled $$$\hat{X}_i=\textbf{D}\alpha_i$$$ at 0.9 mm. To show the improvements of the proposed method, we denoised separately the 1.8 mm and 1.2 mm datasets. The original 1.8 mm and denoised datasets were then upsampled by a factor 2 using both linear spline and cubic spline interpolation as described previously5. We also computed colored fractional anisotropy (FA) maps with a robust tensor estimation15 as implemented in ExploreDTI16.Results
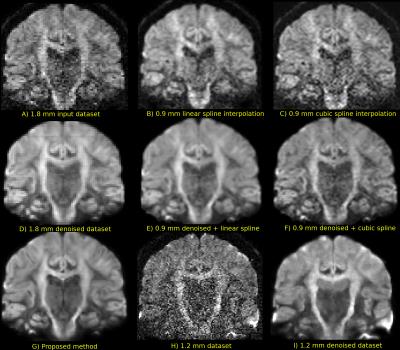
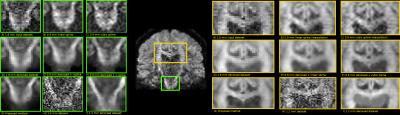
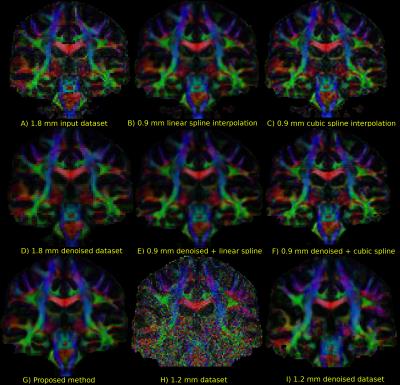
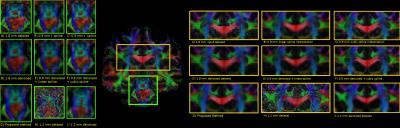
Figure 2 compares the raw data on the 1.8 mm dataset and the various upsampling on the original and denoised datasets on a coronal slice. The 1.2 mm dataset is also shown with its denoised version for anatomical comparison. Figure 3 shows two zoomed regions; the corpus callosum, the cingulum and the corticospinal tract and the pons region with the cerebellum. Figure 4 and 5 show the colored FA maps of the previous figures. Interpolation on the raw data leads to residual artefacts in the interpolated dataset. Applying denoising separately from interpolation introduces line artefacts or blurring, while our proposed combined approach preserves more details, but with the tradeoff of some blocking artefacts. Interpolation results are in agreement with the 1.2 mm raw and denoised dataset, even though they are produced from a 1.8 mm dataset.
Discussion and conclusion
We presented a new framework which combines two usually separate processing steps, namely denoising and upsampling, in a single unified framework. The proposed method links a set of lower and higher resolution patches, which permits reconstruction of dMRI data at a higher resolution than initially acquired without using specialized acquisition schemes. This category of approaches is particularly attractive since they can be applied on any already acquired datasets. As noted previously5, upsampling can reveal finer anatomical details which might help tractography, but care should be taken about possible biases or introduced artefacts in computed metrics such as FA. Moreover, our proposed method could be combined with acquisition techniques1,2,3 to potentially improve the provided anatomical information.
Acknowledgements
Samuel St-Jean is supported by the Fonds de recherche du Québec – Nature et technologies (FRQNT). This research is supported by VIDI Grant 639.072.411 from the Netherlands Organisation for Scientific Research (NWO). Datasets were provided by the Centre d'imagerie moléculaire de Sherbrooke (CIMS) and the Sherbrooke Connectivity Imaging Lab (SCIL), Université de Sherbrooke, Sherbrooke, Québec, Canada.References
1. Ning, L. et al., 2016. A joint compressed-sensing and super-resolution approach for very high-resolution diffusion imaging. NeuroImage
2. Scherrer, B. et al., 2015. Accelerated High Spatial Resolution Diffusion-Weighted Imaging. Information Processing in Medical Imaging
3. Van Steenkiste, G. et al., 2016. Super-resolution reconstruction of diffusion parameters from diffusion-weighted images with different slice orientations. Magnetic Resonance in Medicine
4. Coupé, P. et al., 2013. Collaborative patch-based super-resolution for diffusion-weighted images. NeuroImage
5. Dyrby, T.B. et al., 2014. Interpolation of diffusion weighted imaging datasets. NeuroImage
6. Yap, P.T. et al., 2014. Fiber-driven resolution enhancement of diffusion-weighted images. NeuroImage
7. Manjón, J. V. et al., 2010. Adaptive non-local means denoising of MR images with spatially varying noise levels. J. Magn. Reson. Imaging
8. St-Jean, S. et al., 2016. Non Local Spatial and Angular Matching : Enabling higher spatial resolution diffusion MRI datasets through adaptive denoising. Medical Image Analysis
9. Veraart, J. et al., 2016. Denoising of diffusion MRI using random matrix theory. NeuroImage
10. Rueda, A., et al., 2013. Single-image super-resolution of brain MR images using overcomplete dictionaries. Medical image analysis
11. Mairal, J. et al., 2009. Online Learning for Matrix Factorization and Sparse Coding. The Journal of Machine Learning
12. Glasner, D., et al. 2009. Super-Resolution from a Single Image. International Conference on Computer Vision (ICCV)
13. Koay, C.G., et al, 2009. Probabilistic Identification and Estimation of Noise (PIESNO): A self-consistent approach and its applications in MRI. Journal of Magnetic Resonance
14. Koay, C.G., et al., 2009. A signal transformational framework for breaking the noise floor and its applications in MRI. Journal of magnetic resonance
15. Tax, C. M.W., et al., 2015. REKINDLE: Robust extraction of kurtosis INDices with linear estimation. Magn. Reson. Med.
16. Leemans A. et al., 2009. ExploreDTI : a graphical toolbox for processing, analyzing, and visualizing diffusion MR data. 17th Annual Meeting of Intl. Soc. Mag. Reson. Med.
Figures