3522
Multi-contrast diffeomorphic non-linear registration of orientation density functions1Centre for the Developing Brain, Division of Imaging Sciences and Biomedical Engineering, King's College London, London, UK, London, United Kingdom, 2Florey Institutes of Neuroscience and Mental Health, Melbourne, Australia
Synopsis
Spatial
Introduction
Spatial normalisation of high angular resolution diffusion images (HARDI) is an important prerequisite for group-level analysis of tissue microstructure. Multi-tissue Constrained spherical deconvolution (CSD) is a recently proposed technique to decompose HARDI data into multiple tissue compartments, including cerebrospinal fluid (CSF), grey matter (GM), and white matter (WM) densities, the latter in the form of a full orientation distribution function (ODF)1. Diffeomorphic registration of white matter ODFs has been proposed to provide improved alignment for subsequent group analysis of white matter pathologies2. In this study, we extend this technique by including other non-WM tissue types into the metric driving the registration, and investigate the benefits this provides in terms of overall alignment.Methods
The framework used in this study consists of a symmetric diffeomorphic non-linear registration algorithm adapted to operate on ODFs, taking into the appropriate reorientation of the ODFs required to preserve topology and alignment of oriented features across subjects. The registration proceeds using a multi-resolution pyramid with increasing angular frequency terms. The metric driving the registration consists of the mean squared difference in the spherical harmonics coefficients between the moving and target images, after reorientation (equivalent to the mean squared difference in the ODF amplitudes). The gradient field is smoothed and used to update the displacement field, in an iterative process until convergence is achieved. In this study, we extend the metric by including other tissue types in addition to the WM, and combine them with user-defined relative weights.
We use 20 preprocessed HARDI datasets from the Human Connectome Project (HCP). We estimated three tissue type response functions for WM, grey matter (GM) and CSF for each subject using a data driven method3 which we used to decompose the data into three tissue classes. The images were bias field corrected and intensity normalised.
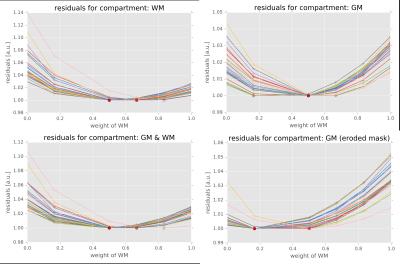
We used this data to assess the influence of WM and GM on the registration accuracy at the single subject level by nonlinearly registering each subject (S1) with a randomly chosen other subject (S2). We subsequently transform S1 into the space of S2 and register this transformed dataset with the original undistorted one. We use the residuals between S1 and the transformed and registered version of S1 to assess the effect of different relative tissue weightings.
Furthermore, we compare group average templates that were generated from those datasets using relative tissue weighting that yielded minimum residuals with a group average template that was generated with only the WM component. Both templates are registered to the common mid-space for visual assessment.
Results
Figure 1 shows the effect of the weightings of the WM component relative to the GM component on the voxel-wise residuals for the inter-subject registration experiment averaged across the brain mask. There is a clear trade-off between between using WM and GM for the alignment of both WM and GM but using both compartments improves the average registration accuracy averaged across the brain. Optimal relative weightings differ, however, depending on which tissue type’s residuals need to be minimised and in which region of the brain is considered.
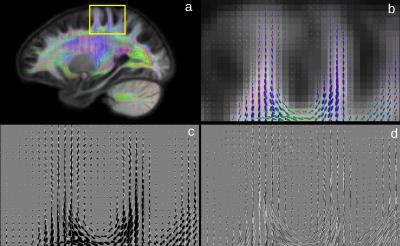
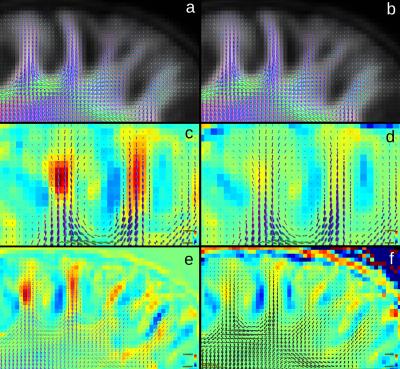
Visual comparison of the WM template generated using only the WM compartment (WMonly) and the template using GM and WM components with equal contribution showed an increased curvature of fibres bending into the cortex (Figure 2) and increased contrast between WM and GM in the cortex (Figure 3).
Discussion and Conclusion
Our results show that including additional non-WM tissue types in the registration metric improves the performance of the registration, as assessed by visual inspection (sharper features), and in simulations (slight reduction in residuals). The optimal weights to assign to each tissue type need to be determined based on the target application, and importance of WM alignment relative to GM alignment. The inclusion of the GM tissue types seems to provide a moderate improvement in the alignment of subcortical white matter features, a feature that may prove beneficial for group analysis of superficial white matter structures such as U-fibres. This may also be of interest in analyses that combine WM and GM information, such as connectomic analysis where cortically-defined regions of interest are used to select streamlines connecting between different regions.Acknowledgements
Data were provided by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University.References
1. Raffelt, D., Tournier, J.-D., Fripp, J., Crozier, S., Connelly, A., Salvado, O., 2011. Symmetric diffeomorphic registration of fibre orientation distributions. Neuroimage 56, 1171–1180.
2. Jeurissen, B., Tournier, J.D., Dhollander, T., Connelly, A., Sijbers, J., 2014. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. Neuroimage 103, 411–426
3. Dhollander T, Raffelt D, Connelly A. Unsupervised 3-tissue response function estimation from single-shell or multi-shell diffusion MR data without a co-registered T1 image. Proc ISMRM Workshop on Breaking the Barriers of Diffusion MRI 2016:5.
Figures