3519
High Spatial Resolution White Matter Fibrography (WMF): Technique Optimization1Boston University, Boston, MA, United States
Synopsis
Purpose: To test the high spatial resolution limits of the white matter fibrography (WMF) technique within the scan time constrains of clinical MRI of about ten minutes total scan time and to optimize the image processing algorithms for rendering the white matter (WM) connectome at the highest level of anatomic detail. Methods: Healthy volunteer was scanned with the quadra-FSE pulse at high spatial resolution in 10min. Results: WMF connectome was constructed using ultra-high b-value (34,000s/mm2) synthetic MRI. Conclusion: High spatial resolution direct rendering of the human brain connectome can be accomplished with a 10min scan.
Purpose
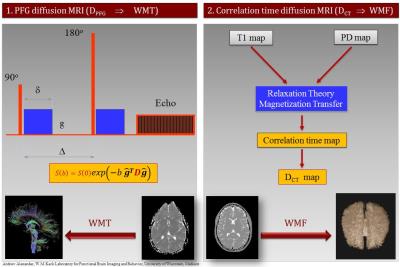
Correlation-time diffusion (DCT) MRI (1) is an alternative diffusion coefficient mapping technique that stems from multi-spectral (MS) qMRI and is highly insensitive to physiologic motion and magnetic susceptibility artifacts because it uses fast spin echo (FSE) pulse sequences. It has been shown recently (2) that the full-brain connectome can be rendered using Synthetic-MRI via DCT-weighting in the high synthetic b-value regime (b>4,000s/mm2); this method was referred to as WM Fibrography in order to distinguish it from diffusion tensor imaging (DTI) WM Tractography, which uses the diffusion encoding via pulsed field gradient (PFG) (Fig. 1) and echo planar imaging (EPI). The purpose of this work was to test the high spatial resolution limits of the WMF technique within the scan time constrains of clinical MRI of about ten minutes total scan time and to optimize the image processing algorithms for rendering the white matter (WM) connectome at the highest level of anatomic detail.Methods
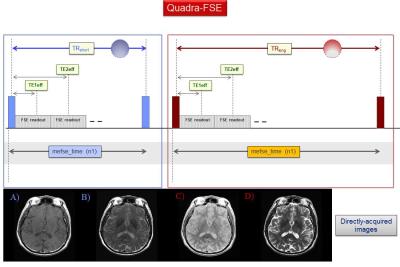
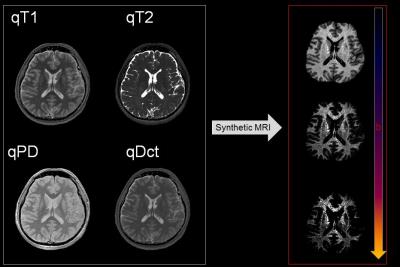
A healthy volunteer (52yo male) was scanned at 3.0T (Discovery MR750w, GE Healthcare, Waukesha, WI) using the quadra-FSE pulse sequence (Fig. 2). This is the concatenation of two dual-echo (DE-) FSE scans differing solely in TR. The acquired images (80 axial slices, 4 images per slice, and 10min total scan time without parallel imaging and NEX=1) were processed with multi-spectral qMRI algorithms programed in Mathcad (version 2001i, PTC, Needham, MA) at the full acquired spatial resolution (0.47 x 0.47 x 2mm3). Spatially coregistered maps of qPD, qT1, qT2, and qDCT were generated for all slices and used as input for the synthetic-MRI algorithm, which has provisions for generating synthetic weightings for all qMRI parameters above. In addition, the algorithm incorporates brain segmentation using a dual clustering algorithm in order to explore the ultrahigh b-value regime; this is necessary in order to avoid the extremely bright subcutaneous fat, which results from its very slow diffusion coefficient (Fig. 3, right column).Results
The qMRI maps were used to generate diffusion-weighted synthetic MR images at b- values ranging the 0-to-50,000s/mm2. Synthetic diffusion weighted images showed a fine, irregular, and clearly defined micro-texture in WM for b>4,000s/mm2. Synthetic DCT-weighted images (Fig. 3, b=34,000s/mm2) were read as a 3D stack in ImageJ (https://imagej.nih.gov/ij/) and resampled to isotropic resolution via bicubic interpolation along the slice-selection direction. The full-brain connectome was rendered using the 3D-viewer plugin (3) in ImageJ leading to the connectome depictions wherein the anatomy of the WM circuitry is visualized directly (Fig. 4). The approximate b value of 34,000s/mm2 was found to provide an optimal compromise between WM texture visualization and SNR.Discussion
White matter tractography (WMT) which derives from the pulsed-field-gradient diffusion-MRI (PFG-dMRI) experiment is the current standard methodology for creating three-dimensional blueprints of the brain’s full circuitry (connectome). Current MRI technologies allow the acquisition of WMT data at a spatial resolution of about 2mm cubic voxels in clinically feasible times (8-15min). Furthermore, WMT techniques estimate indirectly the connectivity patterns between different brain regions from the continuity in the local estimates of fiber direction at each voxel. In contradistinction, WMF provides direct fiber visualization via rendering. WMF does not use PFG diffusion encoding and therefore uses maximally all the imaging gradient power solely for high spatial frequency encoding. WMF has therefore the potential of achieving very high spatial resolution using dedicated ultra-powerful gradient systems as described in the recent literature (4).Conclusion
WMF is a promising complementary alternative to WMT for studying the microarchitecture of white matter, which can generate undistorted high spatial resolution connectomes in clinically feasible (<10min) scan times using standard clinical MRI hardware. All computer algorithms used can be fully automatized and therefore WMF has the potential of being incorporated into routine clinical practice. The use of the quadra-FSE pulse sequences further enhances clinical value because the directly acquired images (T1w, T2w, and PDw) are directly usable for diagnostic interpretation. This work could have implications for the assessment of WM disease and for improving preoperative surgical planning.Acknowledgements
No acknowledgement found.References
1. Jara H. Correlation time diffusion coefficient brain mapping: combined effects of magnetization transfer and water micro-kinetics on T1 relaxation. Proceedings ISMRM (Toronto, Canada) 2008.
2. Jara H, Sakai O, Anderson SW, Soto JA. MR Fibrography: an application of correlation time diffusion synthetic MRI (1.5T and 3.0T). Proceeding RSNA (Chicago) 2016.
3. Schmid B, Schindelin J, Cardona A, Longair M, Heisenberg M. A high-level 3D visualization API for Java and ImageJ. BMC Bioinformatics 2010; 11(1):274.
4. McNab JA, Edlow BL, Witzel T, et al. The Human Connectome project and beyond: Initial applications of 300 mT/m gradients. NeuroImage 2013;80:234-245.
Figures