3504
Compressed sensing diffusion spectrum imaging as a forward-looking alternative to multi-shell diffusion MRI in population imaging1German Center for Neurodegenerative Diseases (DZNE), Bonn, Germany, 2Faculty of Psychology & Neuroscience, Maastricht University, Maastricht, Netherlands, 3University of Bonn, Institute of Computer Science, Bonn, Germany
Synopsis
This study investigates the applicability of two advanced diffusion MRI protocols in population imaging: 3-shell High Angular Resolution Diffusion Imaging (HARDI) and Diffusion Spectrum Imaging accelerated through Compressed Sensing theory (CS-DSI). Group analysis of 20 subjects indicates that CS-DSI performs comparable to 3-shell HARDI in the estimation of microstructure parameters and adds the advantage of high b-value acquisitions, further complimentary biomarkers from the diffusion propagator and a high potential to deliver data well-suited for future developments.
Purpose
To assess
advanced dMRI acquisition techniques for application in high throughput and long-term
population studies.Introduction
Diffusion
MRI (dMRI) provides insights into the microstructural architecture of the brain
white matter and is therefore an important imaging modality for population
imaging. The imaging protocol should deliver reliable data with maximum
potential for future analysis. The well-established 3-shell High Angular
Resolution Diffusion Imaging (HARDI) protocol is the dMRI protocol of choice in
recent population studies.1,2 However, advances in the development
of novel acquisition strategies for fast dMRI acquisition that provide high
resolution of intra-voxel microstructure indicate that Diffusion Spectrum
Imaging3 (DSI) accelerated by the application of the Compressed
Sensing4,5,6 (CS) theory also has high potential to fit this task.
Our work presents the results of a pilot study that was designed based on the
requirements for a long-term population study and conducted to compare a time-matched
3-shell HARDI and a CS-DSI imaging protocol for the acquisition of high-resolution
dMRI data.Methods
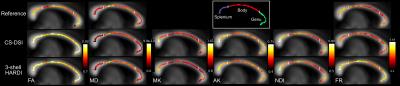
Diffusion MRI scans were acquired from 20 healthy subjects on a 3T Siemens MAGNETOM Prisma scanner (64-channel head-neck coil, 80 mT/m gradient system) using a simultaneous-multi-slice (SMS) dMRI sequence employing threefold slice-acceleration7,8. All protocols applied monopolar diffusion weighting to minimize TE. For each subject, images were collected at 1.5mm isotropic resolution using two advanced imaging protocols: CS-DSI and 3-shell HARDI (Fig. 1). In addition, reference scans were acquired with a standard clinical 2.0mm isotropic DTI protocol in all of the subjects and a dedicated 1.5mm isotropic CHARMED protocol in 4 of the subjects (Fig. 1). The scan time per subject was 11 min for the 3-shell and the CS-DSI protocol, supporting the applicability in population imaging. One minute of additional b=0 scans with reversed phase encoding polarity were collected per protocol.
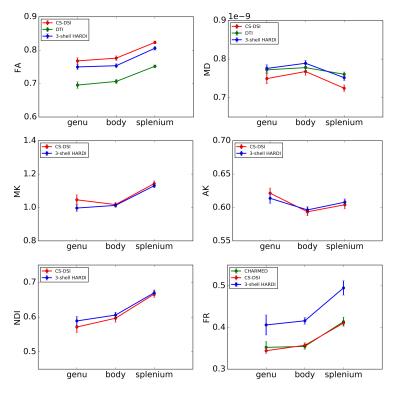
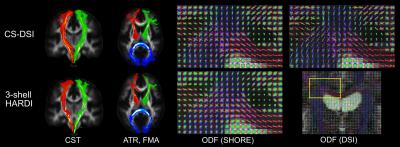
All images are corrected for subject motion and distortions.9,10 CS reconstruction was applied to recover the diffusion propagator from the undersampled DSI acquisitions by means of the discrete Fourier transform combined with a sparsity term.5,6 Using the Maastricht Diffusion Toolbox11 and in-house implementations, several diffusion models were fitted to the data to estimate microstructure parameters: Fractional Anisotropy and Mean Diffusivity (FA, MD; Tensor12 model), Mean and Axial Kurtosis (MK, AK; Kurtosis13 model), Neurite Density Index (NDI; NODDI14), and Fraction of Restricted compartment (FR; CHARMED15). Group analysis was performed using the standard TBSS routine.16 For each parameter, the group mean and standard deviation are subsequently calculated from the mean across all voxels within three regions of interest: the splenium, body and genu of the corpus callosum.11 Furthermore, orientation distribution functions (ODFs) are obtained by means of the SHORE17 model and through direct calculation from the DSI diffusion propagator18. For three white matter tracts, path probability maps are generated by probabilistic tractography.19,20
Results and Discussion
The results presented in Fig. 2 and 3 indicate similar performance of both the CS-DSI and the 3-shell HARDI imaging protocol in the estimation of FA, MD, MK, AK and NDI. FR obtained from CS-DSI, however, is closer to the CHARMED reference than FR from 3-shell HARDI. The upward FA bias of both advanced protocols under investigation compared to the DTI reference is explained by reduced SNR due to longer TE and a smaller voxel size (1.5mm vs. 2.0mm isotropic).21 Fig. 4 visualizes single-subject ODFs and tract-density images overlaid on a white matter atlas22 for three different white matter tracts. Comparable tracking results and SHORE-ODFs are obtained for both CS-DSI and 3-shell HARDI. For CS-DSI, ODFs calculated from the propagator provide superior quality compared to the SHORE-ODFs and allow for potentially better tractography.Conclusion
Our results indicate similar performance of both the 3-shell HARDI and the CS-DSI protocol in the estimation of microstructural parameters and brain connections. One advantage of CS-DSI over 3-shell HARDI is the collection of higher b-value data required for the accurate fitting of specific microstructure models such as the CHARMED model. CS-DSI provides a higher radial resolution than 3-shell HARDI while maintaining high angular resolution and is therefore a more forward-looking acquisition strategy with a greater potential for future developments. Additionally, the diffusion propagator obtained by means of the model-free DSI approach allows for high quality tractography and, potentially, the extraction of further complimentary biomarkers. On the other hand, CS-DSI has slightly smaller SNR than 3-shell HARDI due to higher b-value acquisitions. Future work will investigate if the much larger SNR difference compared to the 2.0mm DTI reference scan may be overcome by denoising or downsampling depending on the application.Acknowledgements
No acknowledgement found.References
1. David C. Van Essen, Stephen M. Smith, Deanna M. Barch, Timothy E.J.
Behrens, Essa Yacoub, Kamil Ugurbil, WU-Minn HCP Consortium. The WU-Minn Human
Connectome Project: An overview. NeuroImage. 2013;80:62-79.
2. Miller KL, Alfaro-Almagro F, Bangerter NK, Thomas DL, Yacoub E, Xu J,
Bartsch AJ, Jbabdi S, Sotiropoulos SN, Andersson JL, Griffanti L, Douaud G,
Okell TW, Weale P, Dragonu I, Garratt S, Hudson S, Collins R, Jenkinson M,
Matthews PM, Smith SM. Multimodal population brain imaging in the UK Biobank
prospective epidemiological study. Nat Neurosc. 2016;19(11):1523-1536.
3. Wedeen VJ, Hagmann P, Tseng WY, Reese TG, Weisskoff RM. Mapping
Complex Tissue Architecture With Diffusion Spectrum Magnetic Resonance Imaging.
Magn
Reson Med. 2005;54(6):1377-86.
4. Menzel MI, Tan ET, Khare K, Sperl JI, King KF, Tao X, Hardy CJ,
Marinelli L. Accelerated Diffusion Spectrum Imaging in the Human Brain Using
Compressed Sensing. Magn Reson Med. 2011;66(5):1226-33.
5. Paquette M, Merlet S, Gilbert G, Deriche R, Descoteaux M. Comparison
of Sampling Strategies and Sparsifying Transforms to Improve Compressed Sensing
Diffusion Spectrum Imaging. Magn Reson Med. 2015;73(1):401-16.
6. Tobisch A, Schultz T, Stirnberg R, Varela G, Knutsson H, Irarrázaval
P, Stoecker T. Comparing Fourier to SHORE Basis Functions for Sparse DSI
Reconstruction. ISMRM. 2015.
7. Setsompop K, Gagoski BA, Polimeni JR, Witzel T,
Wedeen VJ, Wald LL. Blipped-controlled aliasing in parallel imaging for
simultaneous multislice echo planar imaging with reduced g-factor penalty. Magn
Reson Med. 2012;67(5):1210-24.
8. Xu J, Moeller S, Auerbach EJ, Strupp J, Smith SM, Feinberg DA, Yacoub
E, Ugurbil K. Evaluation of slice accelerations using multiband echo planar
imaging at 3T. NeuroImage. 2013;83:991-1001.
9. Andersson JL and Sotiropoulos SN. Non-parametric representation and
prediction of single- and multi-shell diffusion-weighted MRI data using Gaussian
processes. NeuroImage. 2015;122:166-176.
10. Andersson JL, Skare S, Ashburner J. How to correct susceptibility
distortions in spin-echo echo-planar images: application to diffusion tensor
imaging. NeuroImage. 2003;20(2):870-888.
11. Harms R, Goebel R, Roebroeck A. Diffusion microstructure in the
population: variability and effect size of biophysical compartment model
parameters over 100 subjects. ISMRM. 2016.
12. Basser PJ, Mattiello J, LeBihan D. Estimation of the effective
self-diffusion tensor from the NMR spin echo. J Magn Reson. 1994;103(3):247–254.
13. Jensen JH, Helpern JA. MRI
quantification of non-Gaussian water diffusion by kurtosis analysis. NMR Biomed.
2010;23(7):698–710.
14. Zhang H, Schneider T,
Wheeler-Kingshott CA, Alexander DC. NODDI: Practical in vivo neurite
orientation dispersion and density imaging of the human brain. NeuroImage. 2012;61(4):1000-16.
15. Assaf Y, Basser PJ. Composite hindered and restricted model of
diffusion (CHARMED) MR imaging of the human brain. NeuroImage. 2005;27(1):48-58.
16. Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE,
Mackay CE, Watkins KE, Ciccarelli O, Cader MZ, Matthews PM, Behrens TE.
Tract-based spatial statistics: Voxelwise analysis of multi-subject diffusion data.
NeuroImage. 2006;31(4):1487-505.
17. Ozarslan E, Koay C, Shepherd TM, Blackband SJ, Basser PJ, Simple
harmonic oscillator based reconstruction and estimation for three-dimensional
q-space MRI. ISMRM. 2009.
18. Paquette M, Gilbert G, Descoteaux M. Optimal DSI reconstruction
parameter recommendations: Better ODFs and better connectivity. NeuroImage.
2016;142:1-13.
19. Behrens TE, Berg HJ, Jbabdi S, Rushworth MF, Woolrich MW. Probabilistic
diffusion tractography with multiple fibre orientations: What can we gain? Neuroimage.
2007;34(1):144-55.
20. De Groot M, Vernooij MW, Klein S, Ikram MA, Vos FM, Smith SM,
Niessen WJ, Andersson JL. Improving alignment in Tract-based spatial
statistics: Evaluation and optimization of image registration. NeuroImage. 2013;76:400-11.
21. Farrell JA, Landman BA, Jones CK, Smith SA, Prince JL, van Zijl PC, Mori S. Effects of Signal-to-Noise Ratio on the Accuracy and Reproducibility of Diffusion Tensor Imaging–Derived Fractional Anisotropy, Mean Diffusivity, and Principal Eigenvector Measurements at 1.5T. J Magn Reson Imaging. 2007;26(3):756-67.
22. Mori S, Wakana S, Nagae-Poetscher LM, van Zijl PCM. MRI Atlas of Human White Matter. Elsevier, Amsterdam, The Netherlands. 2005.
Figures