3479
The impact of High-Q and High-K on complex fiber structures in the human brain1Brain Imaging and Analysis Center, Duke University Medical Center, Durham, NC, United States, 2Department of Diagnostic Radiology, University of Hong Kong, 3Biomedical Engineering, University of Arizona, Tuscan, AZ, United States
Synopsis
In this diffusion MRI study, we investigated the impact of high angular resolution (high-Q) and high spatial resolution (high-K) on complex fiber structures. It was found that while high-Q was able to resolve crossing fibers within a given region, high-K provided additional spatial details of these crossing fibers in the same location. In addition, diffusion data from high-K improved characterization of high-curvature fibers, which cannot be adequately resolved with high-Q. It is thus concluded that high-K is preferred when both crossing and high-curvature fibers need to be resolved, as in human connectome analysis.
Purpose:
While structural connectome maps of human brain communication are often derived through diffusion tensor imaging (DTI) and fiber tractography, challenges remains to appropriately resolve complex fiber bundles that either cross one another or exhibit high-curvature. In high angular resolution (high-Q) diffusion MRI (dMRI) techniques1, the diffusion behavior within a voxel is observed across multiple tensors to model the behavior of crossing-fibers. Although high-Q data has shown promise in delineating crossing-fibers, the rapid acquisition of dMRI volumes requires spatial resolutions to be as low as 2.0mm. While fewer dMRI volumes can be acquired in a given time when using high spatial resolutions (high-K), the use of sub-millimeter voxels offers an improved resolvability of fibers with high-curvature (u-fibers)2, such as short association fibers connecting adjacent gyri3. In this study, we perform a comprehensive evaluation of the effects of high-Q and high-K sampling on complex fiber structures with both crossing and high-curvature fibers.Methods:
To assess the impact of tractography derived from high-Q and high-K sampling on u-fibers and crossing-fibers, two dMRI data sets were acquired from the same subject with b=800s/mm2. The first data set used high-Q sampling with 90 dMRI directions but at a low spatial resolution with 2.0mm isotropic voxels. This data set was acquired with a 25.6cm FOV in a 128×128 sampling matrix, and a 12sec TR, resulting in an 18.5min scan time. In the second data set, high-K sampling was used to achieve 0.85mm isotropic voxels in 12 dMRI directions using 3D multi-shot EPI data reconstructed with MUSE4. After image reconstruction, motion and eddy current corrections were performed in both data sets using DTIPrep5, and streamline deterministic fiber tractography was performed using first order fiber orientation distribution maps throughout the white matter with MRtrix6.Results and Discussion:
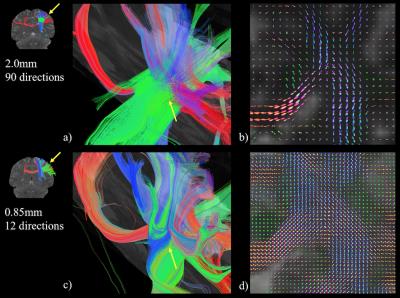
Fig. 1 presents fiber-crossing behavior at the intersection of the corpus callosum (CC), cortico-spinal tract (CST) and superior longitudinal fasciculus (SLF) fiber bundles, shown in red, blue and green respectively. As high-Q dMRI is designed to better delineate crossing-fibers at low spatial resolutions, the CC, CST and SLF fiber bundles in Fig. 1a are shown to pass through one another with clear fiber crossing behavior in the 2.0mm voxels. This behavior is further reflected in the fiber orientation distribution (FOD) map in Fig. 1b, where voxels in a coronal plane of the intersecting region show an equivalent FOD for all three fiber bundles within a single voxel. Rather than modeling crossing-fiber behavior with high-Q, high-K dMRI is designed to physically observe fiber behavior at a finer granularity. When acquired with 0.85mm isotropic voxels (which exhibit a thirteen-fold decrease in voxel volume from 2.0mm voxels), the intersection of the CC, CST and SLF in Fig. 1c does not show the fiber bundles passing through one another, but rather bundles that pass alongside one another before branching to form connections with nearby gyri. This behavior is reflected in Fig. 1d, where the FOD within 0.85mm voxels is predominantly unidirectional at the intersection of the CC, CST and SLF.
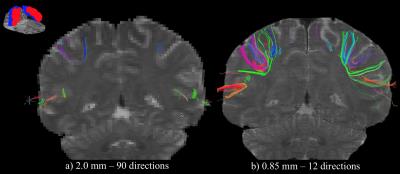
As communication between the sensory and motor cortices is performed through high-curvature fibers that pass through the central sulcus, the behavior of fibers in this region is presented in Fig. 2, where fibers connecting the pre- and post-central gyri are shown. When derived from high-Q data with 2.0mm voxels in Fig. 2a, few short straight fibers form a connection between the adjacent gyri. For high-K data with 0.85mm voxels in Fig. 2b, however, a significant amount of high-curvature fibers are resolved, effectively linking the sensory and motor cortices. In a structural connectome map, fibers derived from 2.0mm data would thus describe a weak connection between these two cortices, irrespective of angular resolution, while a strong connection would result at 0.85mm, even with low angular resolution.
Conclusion:
The design of high-Q dMRI acquisitions at low spatial resolution is to model the behavior of fibers that may cross within the volume of a single voxel. While such a model can effectively delineate fibers that do indeed cross within a voxel, the resolvability of fibers that exhibit curvature within a large voxel is inadequate. With high-K dMRI, the decreased voxel volume has an inherent ability to better observe the true behavior of fiber bundles (both crossing and curving), requiring fewer dMRI directions than in high-Q sampling. Given that high-K data can be acquired in a similar time frame to that of high-Q data (<20min), our data suggests that high-K acquisitions would be preferred when both crossing-fibers and high-curvature fibers need to be resolved, such as in the case of human connectome analysis.Acknowledgements
This work was supported in part by NIH grants NS-075017 and R24-106048.References
1. Tuch, DS, et al. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn. Reson. Med., 2002; 48(4):577-582.
2. Chang, HC, Guhaniyogi, S, Chen, NK. Interleaved diffusion-weighted EPI improved by adaptive partial-fourier and multiband multiplexed sensitivity-encoding reconstruction. Magn. Reson. Med., 2015; 73:1872-1874.
3. Schuez, A., & Miller, R. Cortical Areas: Unity and Diversity. Abington, UK: Taylor & Francis, 2002.
4. Chen NK, Guidon A, Chang HC, Song AW. A robust multi-shot scan strategy fot high-resolution diffusion weighted MRI enabled by multiplexed sensitivity encoding (MUSE). Neuroimage, 2013; 72:41-47.
5. Oquz, I, et al. DTIPrep: quality control for diffusion-weighted images. Neuroinform, 01/30/2014; 8:4.
6. Tournier, JD, Calamante, F, Connelly, A. MRtrix: Diffusion tractography in crossing fiber regions. Int. J. Imag. Syst. Technol., 2012; 22:53-66.
Figures