3454
Providing Ground Truth Quantification of Anisotropic Diffusion MRI Imaging with a Hollow Textile PhantomSudhir Kumar Pathak1, Catherine Fissell1, David Okonkwo2, and Walter Schneider1
1Psychology, University Of Pittsburgh, Pittsburgh, PA, United States, 2Neurosurgery, University Of Pittsburgh Medical Center, PA, United States
Synopsis
A novel Textile Anisotropic Brain Imaging Phantom incorporating textile hollow fibers (taxons with inner/outer diameter 12/34 micron) is used to validate diffusion MRI imaging (dMRI). The taxon intra and extra spaces can be filled with water or deuterium. A taxon crossing pattern and a packing density pattern is manufactured to test orientation and the amount of taxons. NODDI based Intra-cellular volume fraction is correlated with the amount of taxons (r2=0.96) as compared to FA which is poor predictor with r2=0.11. Fiber crossings are estimated using Constrained Spherical Deconvolution techniques and can be resolved for angles greater then 45 degrees.
Introduction
Diffusion reconstruction models and derived anisotropic metrics are used to analyze diffusion images. We are using a new textile based anisotropic phantom [1], the Textile Anisotropic Brain Imaging Phantom (TABIP), provided by an external firm (Psychology Software Tools, Inc). It has a manufactured microstructure of textile hollow fibers called taxons. Taxons are made using polypropylene with an inner/outer radius of 12/34 microns. We test two key hypotheses: 1) the geometrical information estimated from diffusion MRI in a voxel can resolve underlying taxonal crossing, and 2) anisotropic metrics are sensitive to taxon quantity. To test these hypotheses, the TABIP is configured with two different test patterns, crossing patterns for fiber orientation and packing density pattern for the amount of fibers. Packing density pattern is created using 3D-printed chambers with varying density (20%, 40%, 60%, 80%, 100%). Crossing pattern is created in a separate disk with 90, 45 and 30 angles. In this study a spherical harmonic adaptation of generalized q-sampling imaging [2,4] (GQI) is used for the the crossing test. Two quantification methods, Fractional Anisotropy and intra-cellular volume fraction estimated using Neurite Orientation Dispersion and Density Imaging (NODDI [3]) are used for taxonal quantification.Method
TABIP was scanned using a 3T MRI system (TIM Trio, Siemens, Erlangen, Germany) with a 32-channel coil, using a Multi-shell protocol3. The field of view was 240 mm x 240 mm, matrix size 96 x 96, slice thickness 2.4mm (no gap) with 50 slices. The number of gradient directions used for multi-shell was 257, b-values=3000, 5000 sec/mm2, and TR / TE = 9916 ms / 157 ms. In-house MATLAB code implemented the GQI2 based reconstruction of the dODF and (fODF). Fiber tractography is peformed using DSI Studio.Results
Fractional Anisotropy is estimated for each taxon packing density chamber. FA is then plotted against the known number of taxons for each chamber. A correlation coefficient r2=0.11 is estimated between FA and number of taxons (see Figure 2) which shows that FA is a poor predictor of underlying taxonal volume but is sensitive to the voxels with anisotropic water content. The NODDI4 model is used to estimate intra and extra cellular water for each voxel. A correlation coefficient of r2=0.96 is estimated between average intra-cellular volume fraction for each fiber chamber (see Figure 2) and amount of taxons indicating that the NODDI based model is sensitive to the amount of fiber tracts. Fiber ODFs were computed using spherical harmonic adaptation of generalized q-sampling imaging [2] as described in [4]. To evaluate crossing fibers the angle between first two ODF peaks for the sets of voxels in the 90 and 45 degree crossing regions were computed. 90 degree crossings were resolved with mean 81.64 and standard deviation 11.9 degree and 45 degree crossings were resolved with mean 46.6 and standard deviation 4.0 degree in all voxels (see Figure 3). Region with 30 degree crossing is not able to resolved using current reconstruction method.Conclusion
The Textile Anisotropic Brain Imaging Phantom can be used to assess geometrical and anisotropy metrics in dMRI. Diffusion MRI based anisotropic metrics can be sensitive to amount of fibers. Diffusion Tensor derived FA, correlated only at r2= 0.11 with amount of taxons and was not monotonically related to the ground truth number of taxons (see Figure 2). NODDI based intra-cellular volume fraction of water content shows a good correlation with amount of fibers. This suggest that biophysical models like NODDI can be used for quantification of fiber tracts with precise agreement of manufactured ground truth. Advanced diffusion models like spherical harmonic adaption of GQI can resolve crossing more than 45 degree (Figure 3). 30 degree crossing cannot be resolved using this diffusion modeling techniques.Acknowledgements
Contract support US Army W911QY-15-C-0043, VA ECNC F1880References
[1] Catarina et al 2016 [2] Yeh et al 2010 [3] Zhang et al 2012 [4] Pathak et al 2015, ISMRM PosterFigures
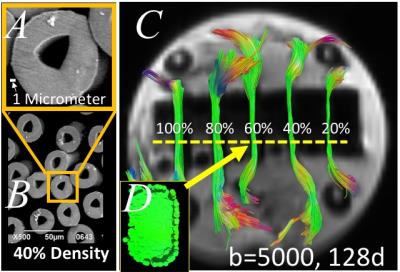
A) Taxon electron microscope image with scale bar,
B) set of taxons
40% packing density; C) Phantom MRI diffusion imaging with overlay T1 with tracts with reducing number of fibers from 100% as 112,640 Taxons; D) MRI of cross section of 60% of
maximum taxon packing density.
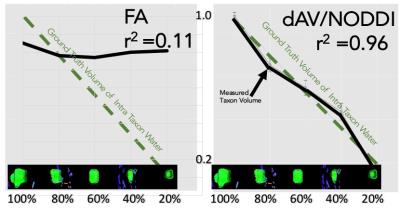
Adapted
a
NODDI (Zhang 2012) FA
shows poor prediction of taxon water whereas NODDI measurement high
correlation.
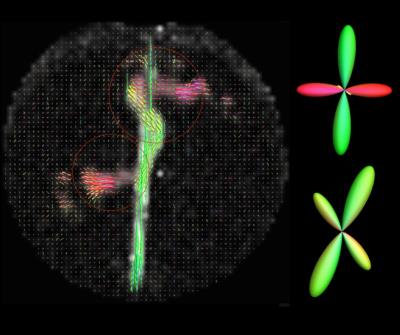
Fiber ODF estimated using
spherical harmonics adapted generalized q-sampling imaging. Fiber crossing is
shown in 90 and 45 degree crossing regions.