3404
Improved algorithm for navigator-based free breathing cardiac diffusion tensor imaging1Siemens Shenzhen Magnetic Resonance Ltd., ShenZhen, People's Republic of China
Synopsis
Cardiac diffusion tensor imaging is an effective way to depict the fiber structure of the myocardium. A navigator(NAV)-based stimulated-echo (STEAM) method was proposed by Nielles-Vallespin to obtain cDTI in vivo. However, its use of a biofeedback process where the subjects had to adapt their breathing pattern in real-time can hinder its clinical implementation. In this abstract, we optimized the NAV accept/reject algorithm, using which the scanning efficiency and the image SNR were both largely improved. Therefore, our work laid a great foundation for the clinical use of free breathing cDTI in the future.
Purpose
The myocardium consists of crossing layers of helical fiber tracts, the integrity of which is fundamental for left ventricle function and could be altered in cardiovascular diseases. Cardiac diffusion tensor imaging (cDTI) is an effective way to depict the fiber structure of myocardium noninvasively1. CDTI is particular challenging, however, because of both cardiac and respiratory motion. A recent work using a stimulated-echo (STEAM) method combined with ECG triggering and perspective navigator (NAV) has shown reproducible results1,2. However, its use of a biofeedback process where the subjects had to adapt their breathing pattern in real-time can hinder its clinical implementation. By improving the NAV accept/reject algorithm, this study aims to increase cDTI’s scanning efficiency and image quality, exploring its potential for clinical use.Methods
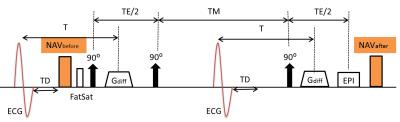
The sequence diagram of ECG-gated diffusion-weighted STEAM with NAVs before and after is shown in Fig.1. In previous work2, the whole sequence continues to run while the acquired data is examined by the NAV accept/reject algorithm of Nielles-Vallespin, where the acquired data will only be accepted if all of the following conditions are satisfied: (1) the position of NAVbefore was in the accept window; (2) the position of NAVafter was also in the accept window; (3) the difference between the position of NAVbefore and NAVafter was in 1mm range.
Our improved algorithm doesn’t require the whole sequence running fulltime (Fig. 2). Rather, after ECG trigger, NAVbefore is performed and condition (1) is first checked. And only when the diaphragm position detected by NAVbefore is within the accept window, then the STEAM module and NAVafter will proceed and conditions (2) and (3) will be checked in the follow-up. If not, however, the process will break and restart from the next ECG trigger (red box in Fig.2).
The experiment was performed on a 3T MR system (MAGNETOM Spectra, Siemens Healthineers, Germany). The cardiac short-axis view of one healthy volunteer was scanned with informed consent. The imaging parameters were: FOV=225×360 mm2, acquisition matrix = 80×128, BW=1955Hz/pixel, slice thickness = 8mm, number of slices = 4, TE = 23ms, diffusion mode = MDDW, diff. direction = 6, b = 350 s/mm2. Either old or new algorithm was used to guide data acquisition in free breathing mode without biofeedback.
Results
Fig.3 shows the respiratory motion of the liver-lung interface. Data in white bar were acquired by NAVbefore and black bar by NAVafter. The green box was the accept window. The red tick indicated that the acquired data was accepted. The real TR in the old algorithm was equal to 2 R-R intervals. While in new algorithm, the real TR was equal to one respiratory cycle (yellow double-headed arrow in Fig.3). One slice of cDWI acquired with old and new NAV accept/reject algorithms were compared in Fig. 4. With new algorithm the total scan time was halved, while the signal-to-noise ratio (SNR) of the image was higher.Discussion and Conclusion
The results demonstrate that the new algorithm improved both the scanning efficiency and the image quality. In the new accept/reject algorithm, the NAVbefore acted like a detector and triggered the STEAM sequence at the beginning of an expiration, which increased the acceptance rate. In addition, the real TR of STEAM sequence was prolonged so the longitudinal magnetization could be fully recovered, and thus led to a better SNR. In all, our new NAV accept/reject algorithm greatly improved the scanning efficiency and the image quality of free breathing cDTI, laying the foundation for its clinical use in the future.Acknowledgements
No acknowledgement found.References
1. Pedro F, Philip J, Laura-Ann M, et al. In vivo cardiovascular magnetic resonance diffusion tensor imaging shows evidence of abnormal myocardial laminar orientations and mobility in hypertrophic cardiomyopathy. JCMR. 2014; 16:87.
2. Nielles-Vallespin S, Mekkaoui C, Gatehouse P, et al. In Vivo Diffusion Tensor MRI of the Human Heart Reproducibility of Breath-Hold and Navigator-Based Approaches. MRM. 2013;70(2):454-465.