3396
Disentangling myelination and axonal density using NODDI and mcDESPOT – a multimodal microstructure imaging approach1Clinical Imaging Sciences Centre, Brighton and Sussex Medical School, Brighton, United Kingdom, 2Neuroimaging, IoPPN, King’s College London, London, United Kingdom, 3Wellcome Trust Consortium for Neuroimmunology of Mood Disorders and Alzheimer’s Disease, London, United Kingdom, 4Centre for Medical Image Computing, University College London, London, United Kingdom
Synopsis
There is an increasing demand for specific tissue microstructure markers that can be related to demyelination and axonal loss. Quantitative MRI techniques are sensitive to microscopic changes but tend to be non-specific. We propose to combine NODDI, a popular MRI diffusion technique, with mcDESPOT, a multi-component relaxation analysis technique, to obtain separate estimates of the volumes of myelin, extra-cellular and intra-cellular spaces. This multimodal MR approach opens the possibility to disentangle changes to myelination and axonal density with the exciting prospect of achieving in vivo histology.
Aim
Develop a multimodal MR approach to quantify myelin and axonal density.Introduction
With growing interest towards g-ratio and white matter plasticity, and the clinical need to understand process of disease, there is an increasing demand for specific tissue microstructural markers that can be related to demyelination and axonal loss. Quantitative MRI techniques such as diffusion MRI, multi-component T2 mapping and magnetization transfer are sensitive to microscopic changes but tend to be non-specific. Neurite orientation dispersion and density imaging (NODDI1) is a popular diffusion MRI technique that estimates indices such as the intra-cellular volume fraction which provides information about neurite density, but such a fraction is relaxation-weighted, and only relative to the sum of intra- and extra-cellular volumes. Multi-component driven equilibrium single pulse observation2 (mcDESPOT) is a multi-component relaxation analysis technique that estimates volume fractions of CSF, myelin and sum of intra/extra-cellular (IE) space. We propose to exploit the complementarity of both techniques in order to obtain separate estimates of the volumes of myelin, extra-cellular and intra-cellular spaces. This opens the possibility to disentangle changes to myelination and axonal density occuring in neurological conditions.Methods
Imaging
We acquired NODDI data and mcDESPOT data for 3 healthy subjects (2 male and 1 female, median age 38, range 27-44 years) using a Siemens Prisma 3T scanner (maximum gradient strength=80mT/m). NODDI images were acquired using TE/TR=95/8200ms, FoV=240x240mm2, matrix size 96x96, slices=60, thickness=2.5mm. Two diffusion shells were used, b=800 and 2000s/mm2 with 30 and 60 non-collinear directions, respectively. McDESPOT imaging protocol included 8 T1-weighted spoiled gradient echo (SPGR) images and 16 balanced T1/T2-weighted steady-state free precession (bSSFP) images acquired using multiple flip angles, as described in 2.
NODDI and mcDESPOT processing
Diffusion data were analysed using the NODDI toolbox to generate
isotropic (vNiso) and intra-cellular (vNic)
volume fractions maps. Then we ran mcDESPOT 3-pool fitting to generate volume
fractions of myelin (vMM), CSF (vMCSF)
and combined intra/extra-cellular space (vMIE). We use
the superscript “N” to indicate maps derived from NODDI, and “M” to indicate
those derived from mcDESPOT. For each participant, NODDI and mcDESPOT maps were
co-registered using SPM12.
Multimodal tissue model
Each voxel may contain CSF and the three separate tissue components: intra-cellular space (I), extra-cellular space (E) and myelin (M), Figure 1. Normalising with respect to the total volume, these fractions must satisfy the condition: $$$V_I+V_E+V_M+V_{CSF}=1$$$.
Mapping volume fractions onto mcDESPOT
The volume fractions extracted from mcDESPOT are vMIE, vMM and vMCSF such that:
$$$v^M_{IE}+v^M_M+v^M_{CSF}=1$$$, $$$v^M_{IE}=V_I+V_E$$$, $$$v^M_M=V_M$$$, and $$$v^M_{CSF}=V_{CSF}$$$
Mapping volume fractions onto NODDI
The volume fractions estimated from NODDI are vNISO and vNIC. It is worth noting that NODDI volume fractions are T2 relaxation weighted such that:
$$$v_{ISO}^N=\frac{V_{CSF}\star e^{-TE/T2_{CSF}}}{V_{CSF}\star e^{-TE/T2_{CSF}}+ V_E\star e^{-TE/T2_{IE}}+ V_I\star e^{-TE/T2_{IE}}}$$$ and $$$v_{IC}^N=\frac{V_{I}\star e^{-TE/T2_{IE}}}{V_I\star e^{-TE/T2_{IE}} + V_E\star e^{-TE/T2_{IE}}}$$$
Assuming T2 is the same for intra-cellular and extra-cellular space2, vNIC will become: $$$v_{IC}^N=\frac{V_{I}}{V_I + V_E}$$$
Calculating volume fractions by combining mcDESPOT and NODDI
Knowing $$$V_I+V_E=v^M_{IE}$$$, we can calculate VI and VE as follows:
$$$V_I=v^N_{IC} \star v^M_{IE}$$$
$$$V_E=v^M_{IE}–V_I$$$
Results and discussion
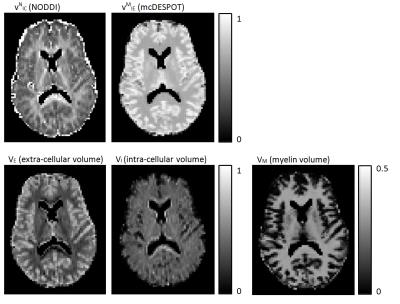
Figure 2 shows voxel-wise maps of the proposed new indices. We can see the intra-cellular volume fraction (VI) looks more uniform across gray and white matter, whereas the extra-cellular volume fraction (VE) shows more contrast between gray and white matter, with lower extra-cellular volume in white matter where myelin takes up significant volume.
Our approach can be seen as complementary to the recently proposed g-ratio framework by Stikov et al.3. Future work will focus on validating this method, with the exciting prospect of allowing in vivo histology to be carried out.
Conclusion
We proposed a multimodal tissue modelling approach by combining different MR modalities NODDI and mcDESPOT. We showed by exploiting both models we can generate more specific tissue properties; particularly separate the intra-cellular and extra-cellular volume fractions. This could help disentangle changes to myelination and axonal density, and possibly provide a significant step towards achieving in vivo histology.Acknowledgements
This work was supported by the Wellcome Trust Neuroimmunology Consortium (NIMA).
Many thanks to Samuel Hurley for providing us with the mcDESPOT sequence.
References
1. Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: Practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 2012; 61:1000-1016.
2. Deoni SC, Matthews L, Kolind SH. One component? Two components? Three? The effect of including a nonexchanging "free" water component in multicomponent driven equilibrium single pulse observation of T1 and T2. Magnetic Resonance in Medicine 2013; 70:147-154.
3. Stikov N, Campbell JSW, Stroh T, Lavelée M, Frey S, Novek J, Nuara S, Ho MK, Bedell BJ, Dougherty RF, Leppert IR, Boudreau M, Narayanan S, Duval T, Cohen-Adad J, Picard PA, Gasecka A, Côté D, Pike GB. In vivo histology of the myelin g-ratio with magnetic resonance imaging. NeuroImage 2015; 118:397-405
Figures