3380
Axon diameter mapping using diffusion MR microscopy embedded in a Monte-Carlo based fingerprint approach1CEA/I²BM/NeuroSpin/UNIRS, Gif-sur-Yvette, France, 2Université Paris-Saclay, Orsay, France, 3CEA/I²BM/NeuroSpin/UNATI, Gif-sur-Yvette, France
Synopsis
In this work, we demonstrate that Monte-Carlo simulations combined with fingerprint approaches can be used to develop decoding tools of the micro-structure using a dictionary learning approach. The validation has been done on a test object mimicking the mid-sagittal plane of a corpus callosum with axon diameters varying according to histological studies. The robustness of the decoding obviously depends on the richness of the dictionary, but, contrary to analytical approaches with highly non linear equations hard to fit practically, such MC approach do not have this kind of limitation, thus opening the way to decode more complex tissue cellular configurations.
Purpose
Diffusion magnetic resonance imaging (dMRI) is a powerful tool to characterize the microstructure of brain white matter, especially the axon diameter/density. Current techniques1,2,3,4 rely on analytical models of the DW signal used to decode the axon diameter/density. Today, models are limited to simple impermeable cylinder geometries because of the high non-linearity of the corresponding equations. To go beyond, we propose an alternative approach based on Monte-Carlo simulations combined with machine learning tools using a dictionary of simulated geometries to decode the microstructure, and we demonstrate that it is as efficient as analytical models to map the axon diameter/density within white matter.Methods
A dedicated framework was developed to assess the feasibility of quantitative axon mapping using a Monte-Carlo approach similar, considering the geometries, to the original ActiveAx1 method proposed in the Camino5 toolkit, eg impermeable parallel cylinders without angular dispersion in the case of this study.
Dictionary of patterns -
63 geometries of impermeable parallel cylinders
with various cylinder diameter (1-10µm) and separated with various
spacings (0 to the cylinder diameter) were designed and saved using
triangulated surfaces (Fig1).
Monte-Carlo
(MC) simulations – the Diffusion Microscopist Simulator
(DMS)6 was used to simulate the Brownian motion
of water molecules within each of the former 63 geometry patterns
(Fig1). For each configuration, 1000
simulations were performed.
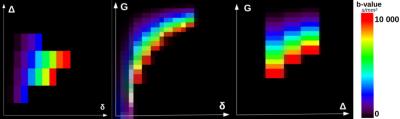
DW signal simulations – the diffusion-decay values corresponding to various settings of a single Pulsed-Gradient Spin Echo sequence were also computed using the DMS tool, but, unlike current ActiveAx protocols (4 different diffusion schemes/shells with specific b-values/diffusion times), and because no scan duration imposes this drastic simplification of the imaging protocol when using simulation, we established a novel strategy based on a Cartesian sampling of the (δ,∆,G) space limited by the MRI system hardware capabilities. For each (δ,∆,G) setting, the diffusion-decay was simulated along 500 diffusion orientations uniformly distributed over the sphere of corresponding b-values. Because we aim at validating our proposed framework on a ex vivo sample of corpus callosum, we used the hardware constraints of a preclinical 11.7T Bruker MRI system, and imposed b (100<= b<=10000s/mm2), G (Geffective<=0,7*Gmax=532mT/m) and TE (<=60ms) constraints. In total, 343 settings stemming from 10δ/4∆/25G/4TE were used (Fig2).
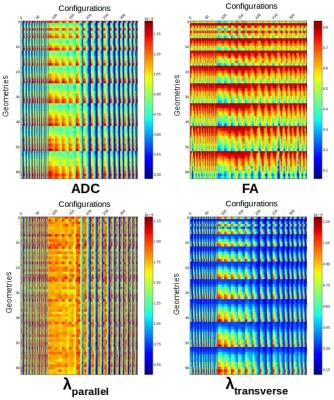
Dictionary of signatures – because the underlying geometry corresponds to homogeneous populations of parallel cylinders, the 500 diffusion decays for each (δ,∆,G) setting could be reduced to their associated DTI coefficients, without loss of information. Of course, for more complex geometries, higher angular models should be used like the SHORE6 decomposition of propagator for instance. This reduction of the problem yielded a resulting matrix corresponding to a dictionary of diffusion-decay signatures for each of the 63 simulated geometries (Fig3).
Decoding of the geometry – this dictionary allowed to construct a decoding tool to automatically recognize the geometry pattern from the observation of its signature established from a collection of DW MR acquisitions. To this aim, a Support Vector Machine7 supervised classifier was trained from the simulated data using the former dictionary.
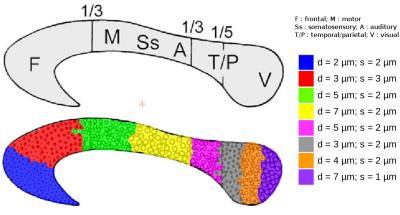
Synthetic
corpus callosum test sample
–
a
test object was created to mimic the mid-sagittal plane of a corpus
callosum (white
matter fibers depicting geometrical configurations close to what was
simulated)
with axon diameters/spacings set
according to
histological
studies8
(Fig4).
Results & Discussion
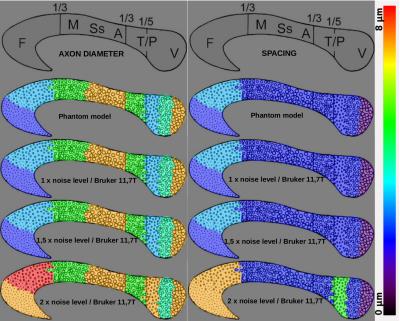
MC simulations were computationally intensive for the 63 geometries (~2 months on a workstation with 40 CPU cores/80 threads with 256GB of memory) but needed to be evaluated only once (Fig1). Figure 2 represents the true domain of available (δ,∆,G) configurations that respects the 11.T Bruker MRI system hardware constraints, the target millimeter resolution constraint and the maximum b, G and TE contraints. The dictionary matrix showing the variation of parameters according to the diffusion sensitization settings and the geometries assessed the feasibility to distinguish the geometry using such information with a supervised classifier (Fig3). As expected, with the designed synthetic corpus callosum (Fig4), the decoding tool was able to recognize the underlying geometry and remains efficient up to twice the standard noise level, the overestimation errors being of the same kind as those found using ActiveAx (Fig5).Conclusion
In this work, we demonstrated that Monte-Carlo simulations are useful to develop decoding tools of the micro-structure using dictionary learning. Their robustness depends on the richness of the dictionary, but, contrary to analytical approaches with highly non linear equations, MC approaches do not have limitation, enabling to simulate/decode more complex configurations, and to extent it to gray matter. Future work will consist of integrating more complex geometries (angular dispersion, bundle crossings, permeability) and to benchmark the approach with respect to analytical approaches.Acknowledgements
This work was partially funded by the European FET Flagship ‘Human Brain Project’ (SP2) FP7-ICT-2013-FET-F/604102References
1. ALEXANDER, Daniel C. A general framework for experiment design in diffusion MRI and its application in measuring direct tissue-microstructure features. Magnetic Resonance in Medicine, 2008, vol. 60, no 2, p. 439-448.
2. ASSAF, Yaniv, BLUMENFELD-KATZIR, Tamar, YOVEL, Yossi, et al. AxCaliber: a method for measuring axon diameter distribution from diffusion MRI. Magnetic Resonance in Medicine, 2008, vol. 59, no 6, p. 1347-1354.
3. ZHANG, Hui, SCHNEIDER, Torben, WHEELER-KINGSHOTT, Claudia A., et al. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage, 2012, vol. 61, no 4, p. 1000-1016.
4. DE SANTIS, Silvia, JONES, Derek K., et ROEBROECK, Alard. Including diffusion time dependence in the extra-axonal space improves in vivo estimates of axonal diameter and density in human white matter. NeuroImage, 2016, vol. 130, p. 91-103.
5. COOK, P. A., BAI, Y., NEDJATI-GILANI, S. K. K. S., et al. Camino: open-source diffusion-MRI reconstruction and processing. In : 14th scientific meeting of the international society for magnetic resonance in medicine. Seattle WA, USA, 2006.
6. YEH, Chun-Hung, SCHMITT, Benoît, LE BIHAN, Denis, et al. Diffusion microscopist simulator: a general Monte Carlo simulation system for diffusion magnetic resonance imaging. PloS one, 2013, vol. 8, no 10, p. e76626.
7. CHANG, Chih-Chung et LIN, Chih-Jen. LIBSVM: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology (TIST), 2011, vol. 2, no 3, p. 27.
8. ABOITIZ, Francisco, SCHEIBEL, Arnold B., FISHER, Robin S., et al. Fiber composition of the human corpus callosum. Brain research, 1992, vol. 598, no 1, p. 143-153.
Figures