3349
To accelerate or not: An Investigation on the Impact of Fast Diffusion Imaging with High Angular Resolution on Diffusion Measures in Fiber Tracts1Department of Computer Science and Information Engineering, National Cheng Kung University, Tainan, Taiwan, 2Department of Radiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, United States, 3Institute of Applied Computer Science, Harvard University, Cambridge, MA, United States, 4Institute of Medical Informatics, National Cheng Kung University, Tainan, Taiwan, 5Graduate Institute of Applied Physics, National Cheng Chi University, Taipei, Taiwan, 6Department of Radiology, Sahlgrenska Academy, Gothenburg University
Synopsis
A novel technique, Fast Diffusion Imaging with High Angular Resolution, is proposed to achieve whole-brain HARDI scans for clinical applications with better geometrical fidelity and shorter scan time. The present study compares tractography results and diffusion properties of each analyzed fiber tract among four-fold segmented (multi-shot) HARDI scans with different acceleration rates and a clinically used sequence with two-fold SENSE. A fully sampled four-shot HARDI scan was used as the reference. The results suggest that the novel acceleration strategy permits a four-minute scan with fairly compatible results while the clinically used method takes ten minutes.
Purpose
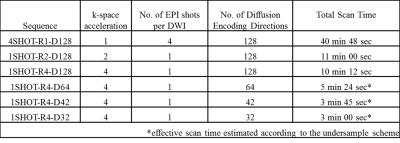
High Angular Resolution Diffusion Imaging (HARDI) provides rich diffusion and fiber crossing information, but scan time has traditionally been too long for practical clinical use. While scan times with HARDI can be reduced to as little as a few minutes, finding the right tradeoff between image quality and speed is not simple. The purpose of this work is a comprehensive analysis of the impact of acceleration on diffusion measures with tractography-derived fiber tracts. The comparison involves a fully sampled multi-shot HARDI with 128 diffusion encoding directions (4SHOT-R1-D128) of 41 minutes as reference standard, a clinically-used single-shot HARDI with two-fold SENSE and 128 directions (1SHOT-R2-D128), and a few different reconstructions with a subset of the fully sampled multi-shot data (four-fold k-space acceleration and varying numbers of diffusion directions 1SHOT-R4 with D128, D64, D42 or D32). The effective scan time of the subsets ranges from 10 to 3 minutes.Methods
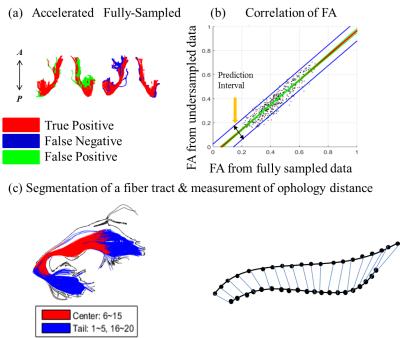
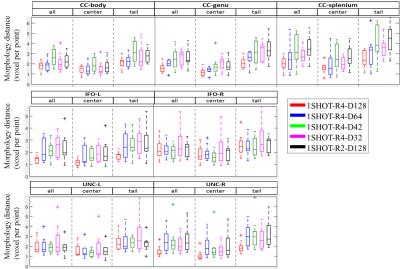
Nine healthy subjects were recruited and scanned following informed consent, using an IRB-approved protocol (3.0T GE Discovery MR750, 50 slices for whole-brain coverage, $$$25\times25$$$ cm2 FOV, $$$2.5\times2.5\times2.5$$$ mm3 isotropic resolution, b = 1500 s/mm2). Two separate scans were performed: (1) Clinically-used single-shot HARDI (1SHOT-R2-D128) with TR = 5000 ms and TE =73 ms, and (2) fully-sampled multi-shot HARDI (4SHOT-R1-D128) with TR = 4500 ms and TE = 64ms. In addition to two b=0 images, the HARDI diffusion encoding scheme was the same in both cases, with encoding directions distributed along a double spiral trajectory. The accelerated sampling schemes were subsampled from the 4SHOT-R1-D128 (see Table 1). Data obtained using the SENSE-accelerated 1SHOT-R2-D128 was reconstructed by the embedded reconstruction engine of the MR system. Otherwise, reconstruction was performed using the previously reported Fast-HARDI1 acceleration and regularization scheme using self-navigation through the multiplexed sensitivity-encoding method2. The reconstruction process is described as: $$\widehat{\rho}_{y,k_{d}}=(F_{d}\Theta^{H}E^{H}_{y}E_{y}\Theta F^{H}_{d}+\lambda M^{-2})^{-1}F_{d}\Theta^{H}E^{H}_{y}(I+\lambda M^{-2})s_{k_{y},d}$$, where $$$s_{k_{y},d}$$$ is the acquired signal,M a diagonal regularization matrix, F a Fourier transform operator and $$$\Theta $$$ a phase correction terms to compensate for motion effects. In the special case of fully-sampled data, the Fourier transformation along d is omitted, and the regularization weighting, $$$\lambda$$$ , is set to be zero. The resulting diffusion weighted images,$$$\widehat{\rho}$$$ , were processed to generate maps of the mean apparent diffusivity (MD), fractional anisotropy (FA), generalized fractional anisotropy (GFA), and orientation density function (ODF), using the Crossing Fiber Angular Resolution of Intra-Voxel structure algorithm3, and a tractography algorithm based on Euler Delta Crossing implemented in the DIPY subroutine4. The b=0 images from different acquisitions were coregistered using SPM85, and the deformation matrices were then used to warp the maps of diffusion measures to the same space for each subject. Three fiber tracts were analyzed in detail: the Corpus Callosum (CC), the Inferio-Fronto-Occipital Fasciculus (IFO) and the Uncinate Fasciculus (UNC). Tract volume, pixel-wise diffusion parameter correlation and averaged morphology distance were computed, to determine how much overlap and deviation could be observed on all measured diffusion parameters, using 4SHOT-R1-D128 as reference standard (see Fig. 1. Only voxels containing more than 3 fibers will be taken as a part of the tract. In the analysis of averaged morphology distance, tracked paths through the tract volumes of the accelerated and reference dataset were paired to find their best counterparts for the estimation of morphology distance based on Euclidean distance after resampling6. The statistics of all the comparisons are presented in terms of medians, quartiles, full range and outliers if the value falls outside 95%, and exhibited on the whiskers plot.Results
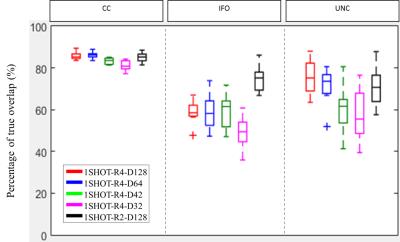
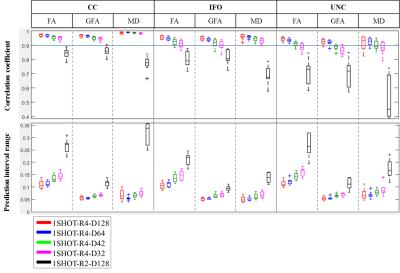
Fig 2 shows the overlapping tract volume fraction among the tested scan scenarios. Over 70% of overlap was observed for1SHOT-R2-D128 while the fraction was greater than 60% for 1SHOT-R4-D42 to 1SHOT-R4-D128. Fig. 3 demonstrates the pixel-wise statistics. The correlation of all diffusion indices from 1SHOT-R4 are all over 90% except those in UNC of 1SHOT-R4-D32, while 1SHOT-R2-D128 shows higher deviation, likely due to difference in geometric distortion. For the averaged morphology distance (see Fig. 4), the medians are all smaller than 4 pixels except for at the tail of CC-splenium by 1SHOT-R2-D128.Discussions and Conclusion
The results suggest that 1SHOT-R4-D128 and 1SHOT-R4-D64 generally have better performances than the clinically used 1SHOT-R2-D128. The 1SHOT-R4-D42 could retain compatible results except for a slightly smaller overlap of tract-derived volume. The difference in tract volume overlap occurs mainly in the ends of the tracts (see Fig 1a). Therefore the present study shows the proposed 4-minute whole brain Fast-HARDI protocol 1SHOT-R4-D42 has the potential to provide fair quality data compatible to the data obtained with 10-minute clinical protocol.Acknowledgements
Support is acknowledged from MOST grant 105-2314-B-006 -044 –MY2, NIH grants R01 CA149342, R21 EB019500, R01 EB010195 and R01 CA160902, and the Mind Research and Imaging Center at the National Cheng-Kung University.References
1. Chao, T.C., et al., Fast diffusion imaging with high angular resolution. Magn Reson Med, 2016.
2. Chen, N.K., et al., A robust multi-shot scan strategy for high-resolution diffusion weighted MRI enabled by multiplexed sensitivity-encoding (MUSE). Neuroimage, 2013. 72: p. 41-7.
3. Landman, B.A., et al., Resolution of crossing fibers with constrained compressed sensing using diffusion tensor MRI. Neuroimage, 2012. 59(3): p. 2175-86.
4. Garyfallidis, E., et al., Dipy, a library for the analysis of diffusion MRI data. Front Neuroinform, 2014. 8: p. 8.
5. Ashburner, J., A fast diffeomorphic image registration algorithm. Neuroimage, 2007. 38(1): p. 95-113.
6. Zimmerman-Moreno, G., et al., Whole brain fiber-based comparison (FBC)-A tool for diffusion tensor imaging-based cohort studies. Hum Brain Mapp, 2016. 37(2): p. 477-90.
Figures