3338
Diffusion imaging with intra volume interleaving of b values1Biomedical Engineering, King's College London, London, United Kingdom, 2Centre for Medical Image Computing, University College London, London, United Kingdom, 3Biomedical Engineering Department, King's College London, London, United Kingdom, 4Centre for the Developing Brain, King's College London, London, United Kingdom, 5Perinatal Imaging, King's College London, London, United Kingdom
Synopsis
Conventional diffusion MRI acquisitions acquire all slices per volume with the same diffusion weighting. This can have two drawbacks: Excessive heating of gradient hardware caused by multiple repeats of the same combination of large drive currents, and low signal at high b-values, which impairs motion correction based on image registration. For placental diffusion MRI, where large slice stack are needed for spatial coverage and anatomical structure can be lost after diffusion, both of these factors can be extreme. We propose intra-volume interleaving of different diffusion weightings, ordered to facilitate image registration for motion correction and minimise gradient heating.
Background
The potential of diffusion MRI (dMRI) to probe microstructural complexity continues to grow with advanced biophysical modelling techniques (1). Emerging applications such as placental dMRI offer fascinating new insights, but also significant challenges, since these advanced analyses require ever more data at high diffusion sensitivity levels (bval) and high angular resolution (HARDI). Furthermore, while frequently employed 2D-single-shot-echo-planar imaging (ssEPI) freezes intra-slice motion, it does not resolve inter-slice movement, so that, especially in non-brain applications, post-processing techniques such as slice-to-volume-reconstruction with registration are required to ensure 3D-continuity.(2,3) In this work we address two problems that are common in dMRI and are accentuated in the placenta application. First, the signal content of high b-value images is strongly attenuated, potentially close to the noise floor. Such images may lack anatomical features, hindering image registration approaches. Second, the required gradients place a high load on the gradient system and require time to cool, which can prolong scan time. A large number of slices, as is required for placental coverage, exacerbates this problem by demanding many EPI shots with unchanging gradient load, which can cause excessive equipment heating. To address both issues, we explore free interleaving of diffusion weighting for each slice within each acquired volume.Methods
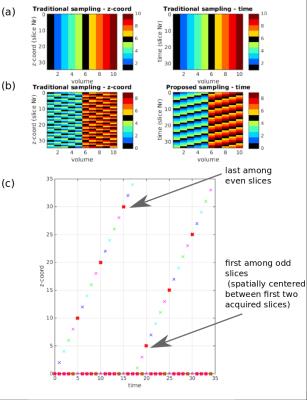
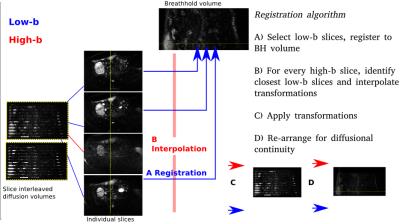
The proposed free diffusion sampling technique breaks the traditional one-volume-one-diffusion-sampling paradigm (Fig.1a) and allows the acquisition of every slice with a different diffusion weighting (Fig.1b). This novel capability, including all required modifications to gradient duty cycle calculations, reconstruction and slice ordering was implemented into the ssEPI sequence on a Philips 3T-Achieva scanner (R3.2 software). An ideal design maximally interleaves low and high b-values for the two above mentioned goals, but ensures that complete volumes are obtained even if the scan is interrupted or abandoned. Therefore, a bloc design with length L was chosen, where L consecutive diffusion samplings were interleaved so that all slices are acquired with all diffusion weightings (i.e. b-value shells) after L volumes. To distribute thermal load the direction of sensitisation can be varied as well as the b value for each slice. The number of required blocks depends on the total number of diffusion samples. To ensure optimal registration properties, the low-b value data was spread out not only maximally in time to densely sample motion patterns but also in space to ensure spatial proximity of every high-b-slice to a low-b-slice. This was realized by maximizing the inter-slice – inter-shot distances. For example, for the frequently used even-odd slice ordering (0-2-4-6….1-3-5-7…), this requires that low b-value-slices are acquired every L shots such that the step stride wraps between slice groups. See Fig.1c for an example with 35 slices, L=5. The required post-processing algorithm was developed in-house using IRTK(4). To ensure optimal registration, a single low-b volume was acquired in a 12s-breath-hold in addition to the diffusion dataset. The complete registration algorithm is shown in Fig.2. Here, the threshold between high-b and low-b was chosen to be b20. Linear interpolation was used between transformations. To illustrate the technique, 4 shell HARDI placental dMRI data with a total of 28 directions (4b0,6b1000,6b2000 and 12b4000), isotropic resolution 2.2mm3, 65 slices, L=5, SENSE1.8 was acquired with a 32ch-cardiac coil. The protocol was repeated with three different acquisition orders (I) conventional slice-stacks with sequentially increasing b-value, (II) optimally ordered volume-interleaving to balance gradient demand and (III) optimally ordered full slice-interleaving.Results
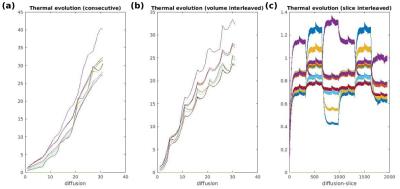
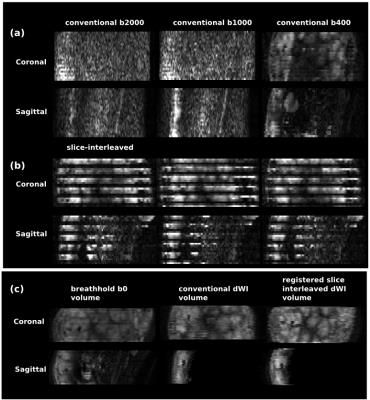
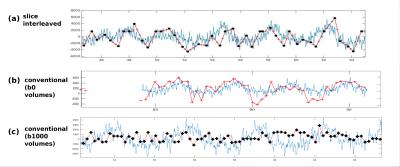
Management of thermal demands (Fig.3) allowed the scan time to be reduced from 10:59s (sequential) to 7:23 (volume-interleaved) to 5:28 (slice-interleaved). Image registration failed for full volumes of high b-value images, but was feasible when temporally adjacent low-b data was available. Figure 4ab illustrates data from conventional and fully interleaved acquisition for 3 consecutive volumes, and Fig. 4c shows registration results with the breathhold target-volume (left) and aligned b400 data for conventional (middle) and the fully interleaved acquisition with the proposed approach (right). The improved alignment is clear. The plotted translations in AP-direction resulting from the low-b registration (Fig.5a overlaid with respiratory belt measurements) nicely depict the breathing cycle, whereas the results from conventional whole-volume approach only accurately depict motion for the b0 volumes (b) but fail for b1000 (c).Discussion
The fully flexible framework presented proved effective in both decreasing acquisition time and enhancing subsequent processing for placental dMRI, but clearly is widely generalizable to any diffusion study that includes high b-values. Future developments could include dynamic field map calculation based on sparse but frequently acquired b0-slices, which can provide significant improvement in the presence of motion or varying B0-fields(e.g. as a result of intestinal gas near the placenta).Acknowledgements
MRC strategic funds (MR/K006355/1), GSTT BRC and NIH-funded Placenta imaging Project: project number 1U01HD087202-01.References
(1)Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage. 2012 Jul 16;61(4):1000-16. doi: 10.1016/j.neuroimage.2012.03.072. Epub 2012 Mar 30
(2) Kuklisova-Murgasova M, Quaghebeur G, Rutherford MA, Hajnal JV, Schnabel JA. Reconstruction of fetal brain MRI with intensity matching and complete outlier removal. Med Image Anal. 2012 Dec;16(8):1550-64. doi: 10.1016/j.media.2012.07.004. Epub 2012 Aug 9.
(3) Ben-Amitay S, Jones DK, Assaf Y. Motion correction and registration of high b-value diffusion weighted images. Magn Reson Med. 2012 Jun;67(6):1694-702. doi: 10.1002/mrm.23186. Epub 2011 Dec 19.
(4) D. Rueckert, L. I. Sonoda, C. Hayes, D. L. G. Hill, M. O. Leach, and D. J. Hawkes. Non-rigid registration using free-form deformations: Application to breast MR images. IEEE Transactions on Medical Imaging, 18(8):712-721, 1999
Figures