Francesco L Palmas1,2, Sarah L Prophet3, Lindsey L Vandergrift3, Taylor L. Fuss3, Shulin Wu3, Chin-Lee Wu3, Adam Feldman3, and Leo L. Cheng3
1Pathology, Massachusetts General Hospital, Charlestown, MA, United States, 2Chemical and Geological Sciences, University of Cagliari, Cagliari, Italy, 3Massachusetts General Hospital, Charlestown, MA, United States
Synopsis
Kidney cancer is the third most common genitourinary
malignancy in the US and leads to over 14,000 deaths and over 61,000 new cancer
diagnoses per year. Since metabolic pathways affect the development of several
malignancies, MRS was applied to highlight the existing alteration between
adjacent benign and cancer tissues. Furthermore, comparison of fresh-frozen paired
samples was performed to assess whether such strategy may interfere and deliver
different results. This approach was able to discriminate according to
pathological condition (benign-tumor) and showed no significant differences in
fresh-frozen pairs.
Introduction
Kidney
cancer is the third most common genitourinary malignancy in the US and leads to
over 14,000 deaths and over 61,000 new cancer diagnoses per year.1 Magnetic
resonance spectroscopy (MRS) has the potential to characterize the metabolic
profiles in tissues and certain biofluids. Since metabolic pathways play a
critical role in the development of several clinical conditions, highlighting
such pathways in renal cell carcinoma (RCC) may help to improve diagnostic
approaches. Indeed, together with ever increasing interests in multi-modality
imaging, MRS applied to onco-metabolomics has shown promising results in
detecting malignancy, predicting tumor stage, and evaluating potential
aggressiveness. The aims of the project are to characterize the metabolic
pattern of RCC specimens and to underline biochemical differences that may help
to distinguish benign from tumor tissues, and to test whether frozen samples
may deliver same results as fresh ones. All are necessary for the discovery of
biomarkers to be translated to in vivo
platforms, and improve clinical practice. Methods
Sixty-seven frozen (48 tumors and 19 adjacent
benign) and 13 fresh-frozen paired kidney biopsy samples were analyzed through intact
tissue MRS. MRS was performed with high-resolution magic angle spinning (HRMAS)
method on a Bruker AVANCE spectrometer operating at 600 MHz. A 4 mm zirconia
rotor was used with Kel-F inserts to generate a 10 μL sample space, and D2O
was added for field locking. Spectra were recorded at 4 ºC with spinning rate at
3600 Hz, and processed using an in-house developed MatLab based program. 58 spectral
regions were identified, normalized by the metabolic region resonance from 0.5 to
4.5 ppm, and evaluated. Results
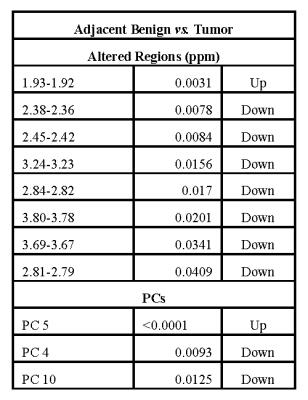
ANOVA
allowed for the separation between adjacent benign tissues and tumor samples highlighting
8 significantly changing regions, while PCA showed 3 statistically different
principal components (Table 1). The tissue
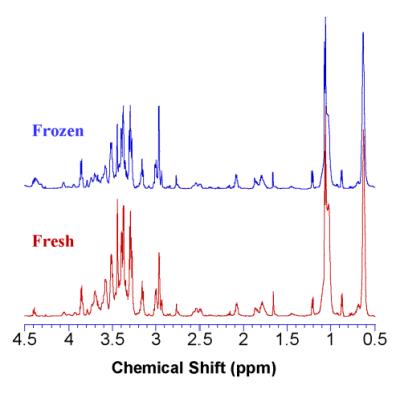
MRS analyses of fresh and frozen samples present no significant difference in
any spectral region (Figure 1). Discussion
Besides the region between 1.93 and 1.92 ppm, a
decrease in spectral intensity is observe for the cancer samples when compared
to adjacent benign ones. This may be explained by higher consumption of the
relative metabolites by the tumor cells. The same trend in noticed for PCs 4
and 10, while 5 seems to be overregulated in benign tissues. The absence of
significant difference between frozen and fresh specimens may be indicative of
appropriate preservation strategies as well as the stability of such tissues that allows for metabolomic
evaluations of previously banked tissue samples. Conclusions
This
preliminary study provides promising results for a further exploration of RCC
characterization through HRMAS MRS technique. The possibility to separate
benign from cancer tissue by their metabolic profile may be translated in in vivo platforms. In turn, this will
help diagnosis in clinical practice, hence allowing for early diagnosis and preventing
potential overtreatment. After MRS, samples will be evaluated with quantitative
histopathology to correlate Furhman and TMN scores with metabolic changes.
Furthermore, a larger sample size will be analyzed to produce a more robust
model.Acknowledgements
Francesco Palmas gratefully acknowledges Sardinia Regional Government
(F.S.E. 2007-2013 UniCa) for the financial support of his PhD scholarship
(P.O.R. Sardegna F.S.E. Operational Program of the Autonomous Region of
Sardinia, European Social Fund 2007– 2013—Axis IV Human Resources, Objective
l.3, Line of Activity l.3.1.). Authors acknowledge support by NIH grant
CA115746 and the A. A. Martinos Center for Biomedical Imaging.References
1.
Siegel RL, Miller RL, Jemal A. Cancer
statistics, 2015. CA Cancer J Clin. 2015; 65(1):5-29.