3252
Analytically-derived Parameter Scouting for Dark-Blood Late Gadolinium Enhancement (DB-LGE) Imaging1Cardiology, BIDMC-Harvard, Boston, MA, United States
Synopsis
In Dark Blood Late Gadolinium Enhanced (DB-LGE) imaging, simultaneous signal nulling of the healthy myocardium and the blood and thus yields superior contrast-to-noise ratio of the scared tissues. The method employs a T2-preparation pulse applied after the inversion recovery pulse to rapidly damp the myocardium magnetization relative to the blood. Accurate timing of the inversion pulse, T2-preparation pulse, and echo acquisition is essential for the success of the technique. In this work, we present a simple method for accurately estimating these parameters through fast low-resolution scouting scans played prior to DB-LGE scans.
Purpose
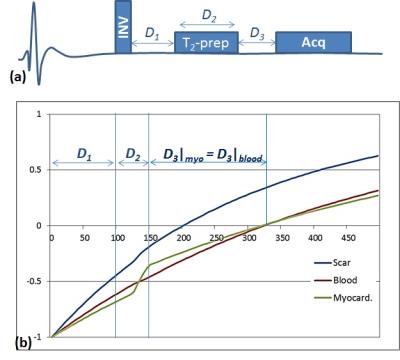
An analytically-derived scouting to determine a proper set of imaging parameters for the DB-LGE sequence is proposed. In DB-LGE, simultaneous signal nulling of the healthy myocardium and the blood can be achieved, which allows producing images of the scared tissues with high contrast-to-noise ratio. In DB-LGE1, a T2-preparation pulse is applied between the inversion-recovery pulse and the image acquisition as shown in Fig 1.a. The T2-preparation is used to temporarily damp the myocardium magnetization relative to that of the blood (utilizing the fact that T2 of the healthy myocardium is much less than that of the blood). This allows a possibility of finding a time point where both the blood and the myocardium signals are nulled as shown in Fig 1.b. Nevertheless, improper selection of the timing parameters, namely D1, D2 and D3, causes the signal of each tissue to be nulled at a different time point as shown in Fig 2.Methodology
Theory: Given arbitrary set of imaging parameters, D1, D2 and D3, the myocardium signal in DB-LGE sequence is given by:
$$M_{myo}(D_1,D_2,D_3)=M_o(1-E_{3myo}+E_{2myo}E_{3myo}-2E_{1myo}E_{2myo}E_{3myo})$$
, where Mo is the fully recovered magnetization, $$$E_{1myo}=e^{-D_1/T_{1myo}}$$$, $$$E_{2myo}=e^{-D_2/T_{2myo}}$$$, $$$E_{3myo}=e^{-D_3/T_{1myo}}$$$.
Equating the above equation to zero, the time D3 that nulls the myocardium can be written as a nonlinear function in D1, and D2; i.e., $$$D_{3myo}=f_{myo}(D_1,D_2)$$$. For the blood signal, a similar equation can be deduced: $$$D_{3blood}=f_{blood}(D_1,D_2)$$$.
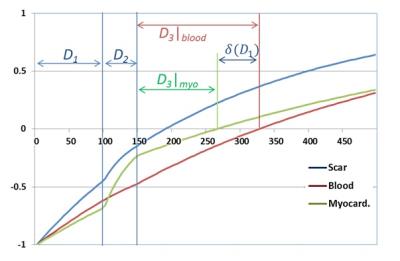
Setting D2 to a small value in order to maintain the highest possible scar signal, then to simultaneously null the blood and the myocardium, one needs to find D1 such that: $$$\delta(D_1)=f_{blood}(D_1)-f_{myo}(D_1)=0$$$. A solution of this equation can be achieved using the iterative Newton-Raphson method:
$$D_{1,n+1}=D_{1,n}-\alpha.\delta(D_{1,n})$$
, where n is the iteration number and $$$\alpha$$$ is the inverse of the partial derivative of $$$\delta$$$ with respect to D1. Instead of calculating $$$\alpha$$$ at each iteration, it is set to a fixed value determined analytically. First, it can be shown that $$$ \alpha\simeq(-\frac{2E_{2blood}}{1+E_{2blood}} +\frac{2E_{2myo}}{1+E_{2myo}})$$$ which is always negative (because T2blood>>T2myo). In fact, calculating the exact value of $$$\alpha$$$ for a wide range of (post-contrast) tissue parameter values reveals that the absolute of its optimal value is always above 3 (this was determined by randomly selecting 10000 different tissue parameter combinations from a pool of all possible (post-contrast) tissue parameter values and calculating for each combination). Therefore, we choose to set $$$\alpha$$$=-3 in all scouting scans. This guarantees convergence to the optimal parameter D1 that yields an image with both the blood and the myocardial signal are null.
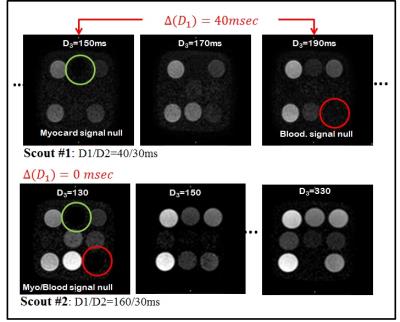
Phantom Experiment: A phantom of nine NiCl2-doped agarose vials with different T1 and T2 values was used to test the developed method. Two vials in the phantom have parameters that mimic those of (post-contrast) healthy myocardium and blood. The scout imaging parameters were as follows: FOV=200×200mm2, voxel size=2×2×8mm3, TR/TE=2.6/1.3ms, bSSFP acquisition, 21 image with D3 varied from 130 to 330ms, with flip angle=55°. In the first scout, D1,0 was intentionally set to a value (= 40ms) that is much smaller than the anticipated correct value (~150). Then, the acquired sequence of scout images is observed to determine the D3 times of nulling the blood and the myocardium, which in turn are subtracted to obtain $$$\delta$$$(D1,0). The iterative equation (above) is then applied to calculate D1,1. The process is then repeated until both the myocardium and the blood are nulled in a single image.
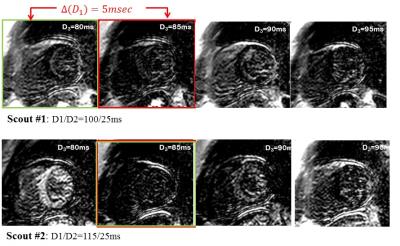
In-vivo Experiment: Scouting for the DB-LGE timing parameters was done on healthy subjects using 1.5T scanner after 35 minutes following Gd-contrast injection. The scouting imaging parameters were: FOV=320×320mm2, voxel size=2×2×8mm3, TR/TE=6.2/3.0ms, flip angle=25°, SENSE rate=2, acq. window=80ms.
Results
Phantom: Figure 3.a shows the first scouting scan of the phantom where the signal of the myocardium and blood vials are nulled at D3=150ms and 190ms, respectively. The second scout is thus performed with D1,1=160ms, where it can be seen in Fig 3b that the signals of the myocardium- and blood- vials are simultaneously suppressed at D3=150ms; that is the optimal timing parameters D1/D2/D3 are 160/30/150ms.
In-vivo: Figure 4a shows the few images of the first scout (with D1/D2=115/25ms). The signals of the myocardium and blood are null at 80ms and 85ms, respectively. The second scout is thus performed with D1=115ms, where it can be seen in Fig 4b that the both the myocardium- and blood- are simultaneously suppressed at D3=85ms; i.e the optimal timing parameters are 115/25/85ms.
Conclusion
A scouting method was presented for determining the timing parameters of the DB-LGE sequence to allow simultaneous nulling of both healthy myocardium and blood pool LGE sequences.Acknowledgements
We thank Gifty Addae, Sophie Berg and Beth Goddu for help with patient recruitment and scanning.References
1. Basha, et al, JCMR 17(suppl 1):O14Figures