3186
Training an Artificial Neural Network by Diffusion-Weighted MRI Data to Differentiate Between Prostate Cancer With High and With Low Gleason Score1Department of Diagnostic, Interventional and Pediatric Radiology, Inselspital, University of Bern, Bern, Switzerland
Synopsis
We prospectively assess the feasibility of using DW-MRI data to train an artificial neural network which distinguishes between prostate cancer lesions with high (≥7) and with low (=6) Gleason scores in 84 patients. The accuracy of the artificial neural network is compared with the accuracy of classification based on apparent diffusion coefficient (ADC) values.
PURPOSE
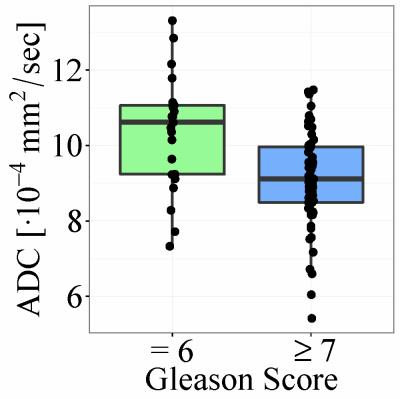
Until recently the vast majority of prostate cancer patients underwent invasive surgery with potential side effects such as impotence or urinary incontinence1. Today, patients with “clinically insignificant” prostate cancer might opt for an “active surveillance” regime during which the lesion is imaged and monitored at regular time intervals. There is no definite consensus on the criteria that define “clinically significant” prostate cancer; however, a commonly accepted definition is a Gleason Score ≥7 upon histopathological analysis of biopsy samples or a volume greater than 0.5 cm3. By fitting a mono-exponential function to the diffusion-weighted MRI (DW-MRI) signal measured within a lesion, it is possible to determine the corresponding Apparent Diffusion Coefficient (ADC). Several studies have suggested a negative correlation between Gleason scores and ADC2,3. However, there is a considerable overlap in ADC values between lesions with high and with low Gleason scores; thus, it does not appear feasible to classify individual prostate cancer patients based on ADC alone4. Other parameters, derived in a nonlinear fashion from the diffusion weighted signal (e.g. by fitting a bi-exponential, stretched-exponential, or kurtosis model), have been found to correlate with Gleason scores5,6. However, it is not clear which of these parameters are best for detecting clinically significant prostate cancer. In this study we assess the feasibility of using DW-MRI data to train an artificial neural network (ANN)7 which distinguishes between lesions with high (≥7) and with low (=6) Gleason scores. The accuracy of the ANN is compared with the accuracy of classification based on ADC values.METHODS
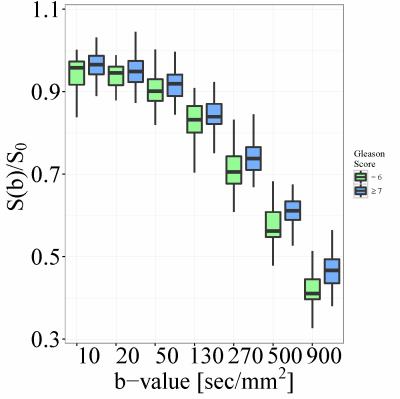
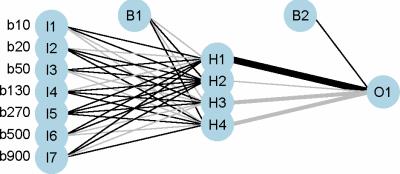
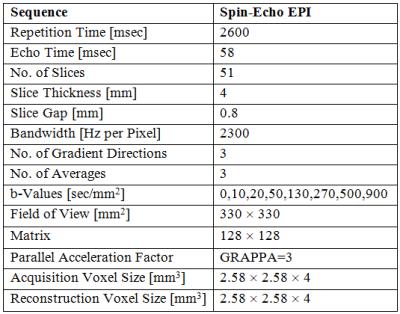
This prospective study includes 84 prostate cancer patients with a Gleason score of 6 or greater and a prostate lesion visible on DW-MRI. These patients were part of a larger cohort of prostate and/or bladder cancer patients scheduled for radical prostatectomy or cystoprostatovesciculectomy. Imaging of the pelvis was performed a median of 12 days (range: 1 to 50 days) before radical prostatectomy and subsequent histopathological analysis; it included coronal 3D T1- and T2-weighted, transverse high-resolution T2-weighted, and transverse DW-MRI sequences (parameters reported in Table 1). A 3T MR scanner (TIM Trio; Siemens Healthcare, Erlangen, Germany), with two phased-array eight-channel coils placed ventrally and on the back, was used for imaging. To reduce artifacts due to peristalsis, patients received 0.5 ml of Glucagon (GlucaGen; Novo Nordisk, Kuesnacht, Switzerland) intravenously before morphological MRI and again 0.5 ml before starting the DW-MRI sequence. Three readers, aware that the patients were scheduled for radical prostatectomy but blinded to any other clinical or histopathological information, independently located the largest prostate cancer lesion on DW-MRI. Any disagreement regarding the lesions’ location was resolved by a fourth reader. The lesions were then delineated in 3D using the RegionGrowingMacro module in MeVisLab (MeVis Medical Solutions AG & Fraunhofer MEVIS, Bremen, Germany). A mono-exponential fit to the DW-MRI data was used to determine the average ADC value within each lesion. The average DW-MRI signal (normalized to S0, 7 b-values) was used as an input to a feed-forward backward-propagation ANN (7 input nodes, 1 hidden layer with 4 nodes, 1 output node) implemented in R 3.3.18,9. Sensitivity, specificity, and accuracy were determined to compare the performance of a classifier based on ADC values and the neural network.RESULTS
Histopathological analysis determined that 23 patients had a Gleason score of 6 and that 61 had a score of 7 or greater. The accuracy of classification based on ADC values was 58/84=69% (sensitivity 42/61=69%, specificity 16/23=70%). The accuracy of the neural network was 77/84=92% (sensitivity 59/61=97%, specificity 18/23=78%). Boxplots of average ADC and DW-MRI data, as well as a schematic plot of the neural network, are presented in Figures 1-3.DISCUSSION
Our study illustrates the feasibility of using ANNs to differentiate between DW-MRI data of prostate lesions with high (≥7) and with low (=6) Gleason scores with an accuracy of 92%. An attempt to classify the same data by combining several diffusion- and perfusion-related DW-MRI parameters in a logistic-regression model did not yield a better classification accuracy than using ADC alone4. An additional advantage of ANNs is that the DW-MRI data can be used directly as an input, eliminating the need for fitting a specific model to the data and thus reducing potential variability due to the employed fitting algorithm10. The main limitation of this study is that we did not cross-validate the accuracy of the neural network on a different dataset; however, additional data is being collected to address this shortcoming.CONCLUSION
ANNs might be used to classify DW-MRI data (acquired at
several b-values) with high accuracy. Nevertheless, validation on additional data is necessary.
Acknowledgements
This study has received funding by the Swiss National Science Foundation (grant 320000-113512); Nano-Tera (RTD: 20NA21_145919); Carigest (Geneva, Switzerland), representing an anonymous donor; Maiores Foundation; Propter Homines Foundation; Kurt and Senta Herrmann Foundation; and Foundation Fürstlicher Kommerzienrat Guido Feger.References
1. Michaelson MD, Cotter SE, Gargollo PC, Zietman AL, Dahl DM, Smith MR (2008) Management of complications of prostate cancer treatment. CA Cancer J Clin 58 (4):196-213. doi:10.3322/CA.2008.0002
2. deSouza NM, Riches SF, Vanas NJ, Morgan VA, Ashley SA, Fisher C, Payne GS, Parker C (2008) Diffusion-weighted magnetic resonance imaging: a potential non-invasive marker of tumour aggressiveness in localized prostate cancer. Clin Radiol 63 (7):774-782. doi:10.1016/j.crad.2008.02.001
3. Turkbey B, Shah VP, Pang Y, Bernardo M, Xu S, Kruecker J, Locklin J, Baccala AA, Jr., Rastinehad AR, Merino MJ, Shih JH, Wood BJ, Pinto PA, Choyke PL (2011) Is apparent diffusion coefficient associated with clinical risk scores for prostate cancers that are visible on 3-T MR images? Radiology 258 (2):488-495. doi:10.1148/radiol.10100667
4. Barbieri S, Bronnimann M, Boxler S, Vermathen P, Thoeny HC (2016) Differentiation of prostate cancer lesions with high and with low Gleason score by diffusion-weighted MRI. Eur Radiol. doi:10.1007/s00330-016-4449-5
5. Zhang YD, Wang Q, Wu CJ, Wang XN, Zhang J, Liu H, Liu XS, Shi HB (2015) The histogram analysis of diffusion-weighted intravoxel incoherent motion (IVIM) imaging for differentiating the gleason grade of prostate cancer. Eur Radiol 25 (4):994-1004. doi:10.1007/s00330-014-3511-4
6. Toivonen J, Merisaari H, Pesola M, Taimen P, Bostrom PJ, Pahikkala T, Aronen HJ, Jambor I (2015) Mathematical models for diffusion-weighted imaging of prostate cancer using b values up to 2000 s/mm(2) : correlation with Gleason score and repeatability of region of interest analysis. Magn Reson Med 74 (4):1116-1124. doi:10.1002/mrm.25482
7. Cheng B, Titterington DM (1994) Neural Networks: A Review from a Statistical Perspective. Statist Sci 9 (1):2-30. doi:10.1214/ss/1177010638
8. R Core Team (2016). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
9. Fritsch S, Guenther F (2016) neuralnet: Training of Neural Networks. https://CRAN.R-project.org/package=neuralnet.
10. Barbieri S, Donati OF, Froehlich JM, Thoeny HC (2016) Impact of the calculation algorithm on biexponential fitting of diffusion-weighted MRI in upper abdominal organs. Magn Reson Med 75 (5):2175-2184. doi:10.1002/mrm.25765
Figures