3147
UNFOLDed spiral SPiRIT TPM enables high spatio-temporal resolution analysis of cardiac function1Dept. of Radiology, Medical Physics, Medical Center – University of Freiburg, Freiburg, Germany, 2Institute of Diagnostic, Interventional and Pediatric Radiology, University Hospital Bern, Bern, Switzerland, 3Cardiology and Angiology, University Heart Center, Freiburg, Germany
Synopsis
A detailed TPM analysis of cardiac function necessitates high spatio-temporal resolution which leads to prolonged scan durations. These scan times are typically too long for data acquisition within a single breath hold. Respiratory navigator-gating can compensate breathing-related motion, but causes an additional increase in measurement time and images with residual motion artifacts. The aim of this study was to combine UNFOLD with variable density spiral SPiRIT TPM to achieve an additional scan time reduction by a factor of 2 and an effective undersampling factor of 6. UNFOLDed SPiRIT TPM demonstrates good results and enables scan times within very short breath holding.
Introduction
MR Tissue Phase Mapping (TPM) is a widely used and powerful approach to assess ventricular function. However, a detailed analysis of the heart wall motion necessitates high spatial and temporal resolution which leads to prolonged scan durations. These scan times are typically too long for data acquisition within a single breath hold (BH) so that navigator gating is applied to compensate for breathing related motion. Unfortunately, navigator-gated measurements are often corrupted by residual motion artifacts and suffer from very low navigator efficiency caused by irregular breathing patterns. Therefore scan time of navigator-gated measurements is typically unpredictable. Alternatively, TPM data could be collected during a single BH which, however, causes a trade-off between spatial and temporal resolution because of the limited acquisition window of 16-20s. Furthermore, patients with cardiovascular diseases often have impaired BH capabilities. The aim of this study was to combine UNFOLD1 with spiral-SPiRIT2 (UNFOLDed spiral SPiRIT) TPM to acquire high spatio-temporal resolution TPM data within 8 heart beats so that only short BH durations are required. Therefore, retrospectively undersampled and UNFOLD-reconstructed velocity encoded spiral datasets of the left ventricle (LV) were compared to the fully sampled data.Methods
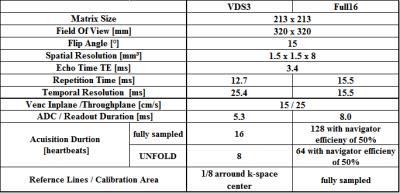
For spiral TPM measurements, a basal short-axis
image of the heart was acquired in 10 healthy volunteers (age 31±5years) on a
3T-Prisma-system (Siemens) using a three-directional velocity-encoded,
black-blood prepared, off-center spiral gradient echo sequence with prospective
ECG gating and 1-1-binomial water excitation. Fully sampled spirals with 16 interleaves (Full16)
were acquired with navigator gating and 3-fold undersampled variable density
spirals with 8 interleaves (VDS3)3 during BH. To investigate the
performance of UNFOLD1 in spiral TPM, both k-space datasets
were retrospectively undersampled by a factor of 2 in time domain in an
interleaved manner. After FFT along the time domain, a low-pass-filter was
applied which sets values outside a 75%-window around the k-f-space center to
zero4. Then, the inverse FFT was applied.
Finally, fully and UNFOLDed datasets were iteratively reconstructed using a CG-SPiRIT
algorithm2,3. Data reconstruction and post-processing
was implemented in Matlab. Post-processing included semi-automatic
segmentation of the LV, eddy current correction and transformation of the
measured in-plane velocities(Vx,Vy) into velocity components perpendicular(Vr) and tangential(Vφ) to the inner heart wall. For segmental analysis, the
basal LV was divided according to the AHA 16-segment model5. Global (averaged over the entire slice)
and segmental systolic and diastolic peak velocities and the corresponding time
to peak (TTP) values were derived for Vr and Vz. Statistical analysis was performed
using a paired student’s t-test to compare full and UNFOLDed data (*p<0.05;**p<0.01).Results
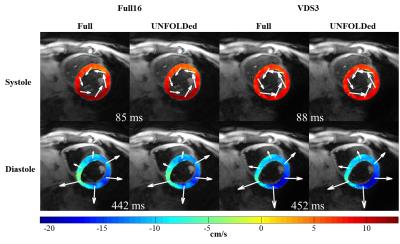
Fully sampled and UNFOLDed undersampled images visually
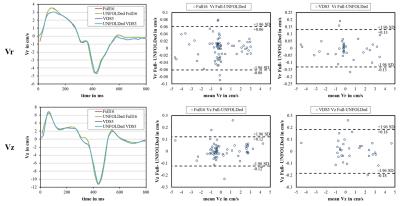
appear in very good agreement (Figure 1). Global velocity time courses of
Vz and Vr were well corresponding (Figure 2). The Bland Altman plots of Vz and
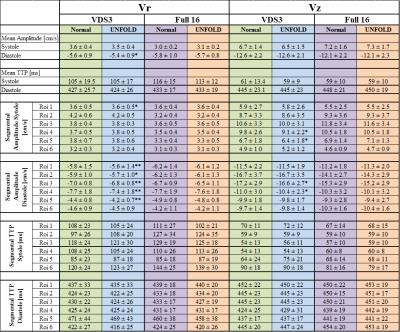
Vr also only show slight deviations between fully sampled and UNFOLDed data(Figure 2). For Full16 acquisition, no differences
were detected for the quantitative values of global and segmental analysis between
full and UNFOLDed datasets (Table 2). Similarly, global and segmental
values for fully sampled and UNFOLDed VDS3 data are in good accordance although
a significant underestimation can be observed for diastolic global and
segmental Vr (Table 2). However, these differences are
small compared to the standard deviation of the associated mean values and
probably negligible in a clinical evaluation.Discussion & Conclusion
We found good agreement of diastolic global and segmental peak velocities and time courses between fully sampled and UNFOLDed spiral TPM data. The slight, but statistically significant underestimation of the Vr peak velocities for VDS3 compared to Full16 might be caused by temporal filtering due to the lower number of timeframes (ratio Full16/VDS=1.6). However, we only employed a simple box-car filter which could be improved with more advanced filter strategies6. A similar approach using spirals in combination with UNFOLD and SENSE reconstruction was already presented by Kowalik et al.7, but achieved a lower spatio-temporal resolution. Further, only global values were presented and not compared to fully sampled datasets as a reference but to self-gated acquisitions. Similarly to our findings, results from Kowalik also showed significant differences in diastolic Vr.
The combination of variable density spiral Spirit TPM with UNFOLD enables an additional scan time reduction by a factor of 2 so that effectively an undersampling factor of 6 is achieved for VDS3 acquisitions. Therefore, TPM acquisitions with high spatio-temporal resolution during very short BH durations (8 heart beats) become possible which might be tolerable even for patients with impaired breath holding. Further work will address optimization of heartrate-dependent filters as well as validation of these results in a larger patient cohort.
Acknowledgements
No acknowledgement found.References
1. Madore B, Glover GH, Pelc NJ. Unaliasing by Fourier-encoding the overlaps using the temporal dimension (UNFOLD), applied to cardiac imaging and fMRI. Magn. Reson. Med. 1999;42:813–828. doi: 10.1002/(SICI)1522-2594(199911)42:5<813::AID-MRM1>3.0.CO;2-S.
2. Lustig M, Pauly JM. SPIRiT: Iterative self-consistent parallel imaging reconstruction from arbitrary k-space. Magn. Reson. Med. 2010;64:457–471. doi: 10.1002/mrm.22428.
3. Menza, Marius, Föll D, Hennig J, Jung B. Spiral SPIRIT Tissue Phase Mapping enables the acquisition of myocardial motion with high temporal and spatial resolution during breath-hold. In: Proceedings of the 24th Annual Meeting of ISMRM. Singapore; 2016. p. 3131.
4. Tsao J. On the UNFOLD method. Magn. Reson. Med. 2002;47:202–207. doi: 10.1002/mrm.10024.
5. Imaging AHAWG on MS and R for C, Cerqueira MD, Weissman NJ, et al. Standardized Myocardial Segmentation and Nomenclature for Tomographic Imaging of the Heart. Circulation 2002;105:539–542. doi: 10.1161/hc0402.102975.
6. Kellman P, Sorger JM, Epstein FH, McVeigh ER. Low Latency Temporal Filter Design for Real-Time MRI Using UNFOLD. Magn. Reson. Med. Off. J. Soc. Magn. Reson. Med. Soc. Magn. Reson. Med. 2000;44:933–939.
7. Kowalik GT, Muthurangu V, Khushnood A, Steeden JA. Rapid breath-hold assessment of myocardial velocities using spiral UNFOLD-ed SENSE tissue phase mapping. J. Magn. Reson. Imaging 2016:n/a-n/a. doi: 10.1002/jmri.25218.
Figures