3135
Free-breathing non-contrast 3D radial respiratory motion-resolved versus motion-corrected whole-heart coronary MRA at 3T1Department of Radiology, Hospital Lausanne (CHUV) and University of Lausanne (UNIL), Lausanne, Switzerland, 2Center for Biomedical Imaging, Lausanne, Switzerland, 3Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland
Synopsis
Coronary MRA performed using respiratory self-navigation techniques that apply retrospective motion-correction suffer from artifacts originating from static anatomical structures surrounding the heart. This becomes even more complicated at 3T due to insufficient fat suppression. Here we implemented free-breathing self-navigated 3D coronary MRA at 3T. Instead of using a motion model, we resolved the motion using 4D k-t sparse SENSE. Resulting images were then quantitatively compared with those reconstructed from the same datasets but using a 1D motion correction. We demonstrate that non-contrast whole-heart coronary MRA can be performed at 3T and that 4D motion-resolved reconstruction effectively minimizes adverse effects of respiratory-motion.
Purpose
Coronary MRA is commonly performed during free-breathing and using navigator-gated acquisitions. However, disadvantages of navigators include unpredictable scanning times and operator dependency. To overcome this, self-navigation techniques with retrospective respiratory motion correction have been developed [1,2]. Unfortunately, such correction may lead to artifacts that originate from static anatomical structures surrounding the heart, and this problem may be exacerbated at 3T where fat signal may not always be homogeneously suppressed in the entire field of view. As an alternative to motion correction, respiratory motion-resolved reconstruction using k-t sparse SENSE [3] has recently been introduced [4]. Since this approach obviates the need for correction, and on the above background, we posit that motion resolved reconstruction leads to improved image quality at 3T when compared to that obtained with motion correction. To test this hypothesis, we implemented a free-breathing self-navigated 3D golden angle radial gradient echo (GRE) imaging sequence using water excitation for fat signal suppression at 3T [5]. The acquired image data were then reconstructed with 1D motion correction on the one hand, and with respiratory motion-resolved 4D k-t sparse SENSE on the other. The resultant images were then quantitatively compared.Methods
Non-contrast enhanced coronary MRA was performed in (n=7) healthy
adult volunteers on a 3T clinical scanner (Magnetom PRISMA, Siemens
Healthcare). Data were acquired using a prototype ECG-triggered respiratory self-navigated
free-breathing 3D radial GRE imaging sequence preceded by an adiabatic T2
preparation module (T2-Prep) to improve blood tissue contrast [6]. Off-resonant
water excitation (WE) as described previously [5] was used as a lipid-nulling strategy
to enhance contrast at the level of the coronary arteries and to minimize image
artifacts originating from background tissue. 3D volumes with an isotropic voxel
size of 1.1 mm3 were acquired with the following imaging
parameters: field of view (220 mm)3, matrix size 1923, TET2-Prep
= 40 ms, RF excitation angle 18°, TE/TR = 2.5 ms/5.6 ms, with 24 radial readouts
per segment for a total of ~15k k-space lines. All datasets were 1D respiratory
motion-corrected using a superior-inferior (SI) projection acquired every 24
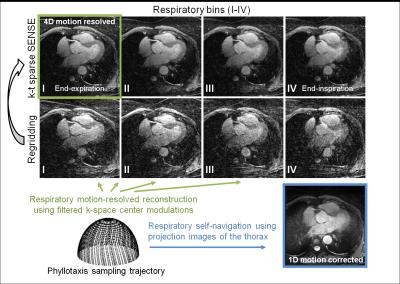
k-space lines as previously described [2]. The same data also underwent motion-resolved
4D (x-y-z-respiratory) k-t
sparse SENSE reconstruction [4]. For this, respiratory
signals were extracted from the filtered k-space center modulations and used to
sort all k-space readouts in 4 different respiratory bins (Fig. 1), ranging from end-expiration to end-inspiration. Respiratory motion-resolved
images were then obtained by solving:
$$\underset{m}{\arg\min}\parallel F\cdot C \cdot m - s \parallel ^2 _2 + \lambda _1 \parallel D_1m \parallel _1 \quad , $$
where F represents the non-uniform fast Fourier transform (NUFFT)
operator, C the coil sensitivity
maps, m the 4D image data set to
reconstruct (where the 4th dimension is the respiratory dimension), s the radial k-space data, D1 the finite difference
operators applied along the respiratory dimension, and λ1 = 0.05 a regularization parameter that was empirically
determined. Both the motion-corrected and motion-resolved
images were qualitatively compared by visual inspection. For quantitative
comparison, vessel
sharpness of the first 4 cm of both the right coronary artery (RCA) and left
anterior descending (LAD), and maximum visible length of both coronaries were
calculated using SoapBubble [7]. Statistics were computed via two-tailed
student’s t-test for paired data.
Results and Discussion
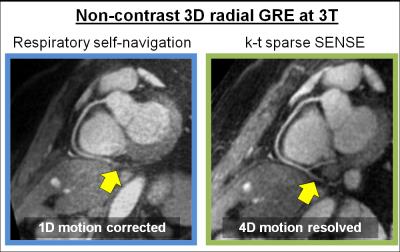
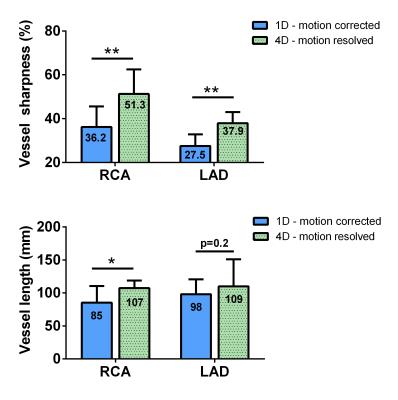
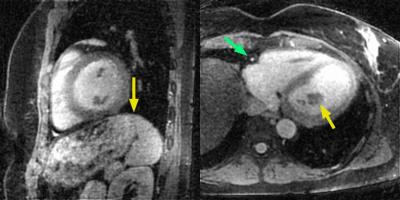
Volunteer data were successfully acquired during free-breathing scans and reconstructed using both motion-compensation methods. The vessel sharpness improved from 36.2% to 55.2% (p=0.004) and from 27.5% to 38.6% (p=0.003) for the RCA and LAD, respectively (Fig. 3a). The length of the vessel that could be tracked improved from 85 mm to 107 mm (p=0.04) and 98 to 109 mm (p=0.2) for RCA and LAD respectively (Fig. 3b). These results obtained using GRE at 3T without contrast agent are consistent with the findings from a recent study using bSSFP at 1.5T [4] - and this suggests that respiratory motion-resolved image reconstruction may be a valuable alternative to self-navigation. When comparing the 4D motion-resolved versus the 1D motion-corrected data, visual inspection demonstrated enhanced anatomical detail. The improved image quality in 4D motion-resolved images may be related to the complex respiratory-induced 3D motion of the heart, which is taken into account to some extent by using the motion-resolved reconstruction (Fig. 1, and Fig. 4 as animated GIF online).Conclusion
Non-contrast enhanced isotropic 3D radial GRE coronary MRA was performed at 3T and motion-resolved sparse reconstruction efficiently minimizes adverse effects of respiratory motion. This leads to significantly improved visualization of the coronary arteries.Acknowledgements
The authors would like to acknowledge the the CIBM, and funding obtained from NanoteraReferences
[1] Stehning et al., Magn Reson Med 54, 476-480 (2005)
[2] Piccini et al., Magn Reson Med 68, 571-579 (2012)
[3] Feng et al., Magn Reson Med, doi: 10.1002/mrm.25665 (2015)
[4] Piccini et al., Magn Reson Med, In press (2016)
[5] Bastiaansen et al., Proc Intl Soc Mag Reson Med 24, 1072 (2016)
[6] Nezafat et al., Magn Reson Med 55, 858-864 (2006)
[7] Etienne et al., Magn Reson Med 48, 658-666 (2002)
Figures