3022
Automated Voxel Placement: A Linux-based Suite of Tools for Accurate and Reliable Single Voxel Coregistration1Psychiatry and Behavioral Neurosciences, Wayne State University School of Medicine, Detroit, MI, United States
Synopsis
Single-voxel magnetic resonance spectroscopy (MRS) provides quantification of brain metabolite levels in vivo. However, MRS studies suffer from an often overlooked source of error variance: inconsistent voxel placement. It is well-established that metabolite levels vary by brain region and voxel tissue composition. Thus, inconsistent voxel placement increases likelihood of Type I and II errors. To address this problem, we developed and evaluated a novel and automated method of prescribing voxel placements at the time of scanning. Results demonstrated a significant improvement in prescribing accurate and reliable voxel placements between and within subjects compared to manual placement and published methods.
PURPOSE:
Single-voxel magnetic resonance spectroscopy (MRS) is a powerful technique that provides quantification of brain metabolite levels in vivo. However, MRS research studies suffer from an often overlooked and unreported source of variance: inconsistent voxel placement between subjects and within subjects. It is well-established that metabolite levels vary across brain regions and by voxel tissue composition (percentage gray vs. white matter)1-3. Inconsistent voxel placement increases the likelihood of Type I and II errors in MRS research studies. Several excellent automated procedures exist for prescribing voxel replacement within a single subject across time (within-subject reliability: 97.8%)4. These applications are well-suited for clinical applications (e.g. tracking medication response). However, to the knowledge of the authors, there is only one published method4 for automated voxel placement across subjects, which exhibited moderate voxel placement accuracy (overlap between each subject voxel and a standard reference voxel: 81%-84%)4. Between-subject reliability (geometric voxel overlap across all subjects) was not reported in that study, but the literature indicated that manual placement (most commonly used approach) was associated with ~70% overlap5. The purpose of this study was to develop and evaluate an automated procedure for accurate and reliable single-voxel prescription.METHODS:
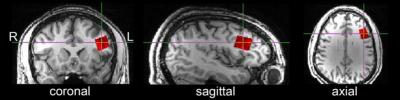
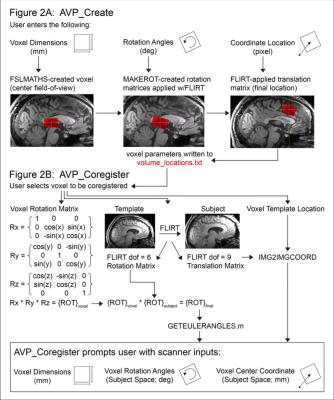
The automated voxel placement (AVP) suite consists of three Linux-based scripts utilizing FSL, FreeSurfer, and Matlab: ‘avp_create’, ‘avp_coregister’, and ‘avp_overlap’. ‘Avp_create’ allows the user to create a library of voxels in a template image for future coregistration (Figure 2A). At the scanner, the user selects a voxel for coregistration. ‘Avp_coregister’ will coregister the voxel in ‘real-time’ (processing time: ~2 min) to subject space based on the acquired T1-weighted images, and output values required to prescribe the voxel (Figure 2B). Finally, ‘avp_overlap’ quantifies geometric voxel overlap between- and within-subjects, and voxel tissue composition (i.e. percentage gray and white matter). Participants (N=13) were recruited locally and provided informed consent. Two voxel placement procedures were evaluated: manual placement (n=5) and AVP (n=8). A subset of AVP participants (n=4) completed a follow-up scan. All imaging was conducted on a 3T Siemens Verio system with 32-channel receive-only head coil. High-resolution T1-weighted structural scans were collected using the 3D Magnetization Prepared Rapid Gradient Echo (MPRAGE) sequence with the following parameters: TR=2.2s, TE=3ms, TI=799ms, flip angle=13°, Field-of-View (FOV) = 256x256x160mm and 1mm3 pixel resolution. The voxel placement evaluated in this study is depicted in Figure 1. Voxel placement accuracy and reliability were evaluated using ‘avp_overlap’. Three sets of analyses were conducted: 1) percentage of geometric overlap between each subject’s voxel and the reference voxel (i.e. accuracy), 2) percentage of geometric overlap shared among all subjects at each time point (i.e. between-subject reliability), and 3) percentage of geometric overlap and tissue voxel composition consistency [coefficient of variation (CV)] within each subject across the two time points (i.e. within-subject reliability). One-way ANOVAs were used for significance testing.RESULTS:
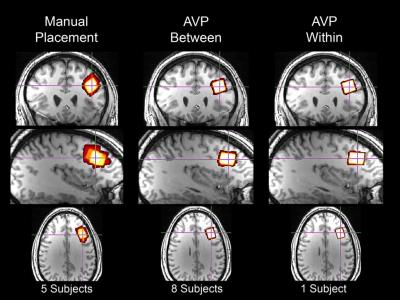
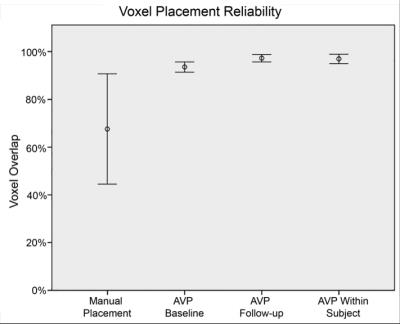
The modal participant was a 28-year-old (range=21-34yrs) African-American (76.9%) male (84.6%). AVP was more accurate than manual voxel placement at baseline (n=8) and follow-up (n=4): F(1,12)=12.84, p<.005 and F(1,8)=6.76, p<.05, respectively. Mean accuracy (± 1 standard deviation [SD]) using AVP was higher [96.2% (2.95%) at baseline and 97.6% (1.41%) at follow-up] than manual voxel placement [67.7% (22.8%)]. Median accuracy using AVP was substantially higher (Cohen’s d ≥ 2.3; very large effect) than most accurate published method: 97.9% vs. 81%-84%4. Within-subject reliability was evaluated two ways: voxel overlap and voxel tissue composition. Mean (± 1 SD) within-subject overlap using AVP was 97.3% (2.1%) (Figures 3,4). Median within-subject overlap using AVP was comparable to the most reliable published method: 97.9% vs. 97.8%, respectively4. Mean gray and white matter voxel composition percentage were: 32.2% (range=20.4%-38.3%; CV=10.8%) and 65.5% (range=58.5%-79.5%; CV=5.2%), respectively. Between-subject reliability was quantified as the percentage of voxel overlap shared across all subjects at each time point (Figure 3). Mean (±1 SD) between-subject overlap using AVP [94.2% (1.0%) at baseline and 97.2% (0.5%) at follow-up] was higher than manual placement [67.7% (20.4%); a value that is consistent with the literature]5 (Figure 4).DISCUSSION:
Inaccurate and inconsistent prescription of MRS voxels is an often overlooked source of error variance in single-voxel MRS studies. In the present study, we developed and evaluated an automated approach of prescribing voxel placements at the time of scanning. Results from this study demonstrated that AVP was more accurate and reliable than manual voxel placement (most common approach used in MRS research) and the most accurate published approach in the literature4.CONCLUSION:
The AVP suite provides accurate and reliable automated single-voxel placement prescription using an a priori template-driven approach.Acknowledgements
Funding generously provided by NIDA (F31 DA040369; awarded to EAW), Wayne State University School of Medicine (New Investigator Grant; awarded to EAW), State of Michigan (Young-Lycaki funds), and the Detroit Wayne Mental Health Authority.
The authors thank Caroline Zajac-Benitez, Michael Lisieski, Natalie Wiseman, Brian Silverstein, Vikas Kodak, Andrew Neff, Chaitali Anand, and Wafaa Sweidan for their assistance.
References
1. O'Neill J, Schuff N, Marks WJ, Feiwell R, Aminoff MJ, Weiner MW. Quantitative 1H magnetic resonance spectroscopy and MRI of Parkinson's disease. Movement disorders 2002;17(5):917-927.
2. Brex P, Parker G, Leary S, Molyneux P, Barker G, Davie C, Thompson A, Miller D. Lesion heterogeneity in multiple sclerosis: a study of the relations between appearances on T1 weighted images, T1 relaxation times, and metabolite concentrations. Journal of Neurology, Neurosurgery & Psychiatry 2000;68(5):627-632.
3. Block W, Träber F, Kuhl C, Fric M, Keller E, Lamerichs R, Rink H, Möller H, Schild H. 1H-MR-spektroskopische Bildgebung bei Patienten mit klinisch gesichertem Morbus Alzheimer. 1995. Thieme. p 230-237.
4. Storrs J. Automatic Real-Time Targeting of Single-Voxel Magnetic Resonance Spectroscopy: University of Cincinnati; 2010.
5. Dou W, Speck O, Benner T, Kaufmann J, Li M, Zhong K, Walter M. Automatic voxel positioning for MRS at 7 T. Magnetic Resonance Materials in Physics, Biology and Medicine 2015;28(3):259-270.
Figures