3013
MASE-sLASER, a short TE matched chemical shift displacement error sequence for single voxel spectroscopy at ultrahigh field1Erwin L. Hahn Institute for Magnetic Resonance Imaging, Essen, Germany, 2Translational and Molecular Imaging Institute, New York, NY, United States, 3Donders Institute for Brain, Cognition and Behaviour, Nijmegen, Netherlands
Synopsis
Conventional sLASER sequence has different Chemical Shift Displacement Error (CSDE) in one of the three slice selection directions. In this work, a short TE matched CSDE sLASER sequence (MASE-sLASER) has been implemented using the novel MASE pulses for single voxel spectroscopy at 7T. The matched low CSDE of this sequence in all three directions provides more exact representation of the metabolites in the imaged voxel. The short duration of the MASE pulses with acceptable bandwidths have made it possible to achieve a TE as short as 28 ms for the MASE-sLASER sequence despite having one more RF pulse than the conventional sLASER sequence.
Purpose
In this work, a short TE matched chemical shift displacement error (CSDE) sLASER1 sequence (MASE-sLASER) is implemented using the novel MASE2 pulses.Methods
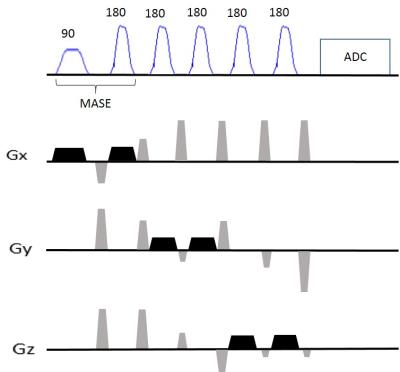
The MASE-sLASER sequence (Figure1) has been implemented using MASE for slice selection in one direction and two pairs of adiabatic SLR refocusing MASE pulses for slice selection in the two other directions. A 5 ms SLR excitation pulse with 3.52 KHz bandwidth and a 2.5 ms adiabatic SLR refocusing pulse with 3.24 KHz bandwidth were used to excite and refocus the signal from the first slice. Two pairs of the same adiabatic SLR refocusing pulses with 2.5 ms duration and 3.24 KHz bandwidth were used for slice selection in the two other directions. In vivo scans were performed on a 7T system (Magnetom 7T, SIEMENS Healthcare GmbH, Germany) with 32 channel Rx and 1 channel Tx head coil (Nova Medical, NY). Two healthy subjects participated with ethical approval from institutional ethics committee. Anatomical reference was acquired using 3D MPRAGE. B0 shimming was performed using FASTESTMAP3. Single voxel MRS data were collected at short TE=28 ms from a 20x20x20 mm3 voxel positioned at occipital region using MASE-sLASER sequence and at TE = 38 ms from the same voxel using sLASER sequence (TR=4500ms, averages=32, scan time=2:42 mins for both sequences). Data were analyzed using JMRUI software4.Results
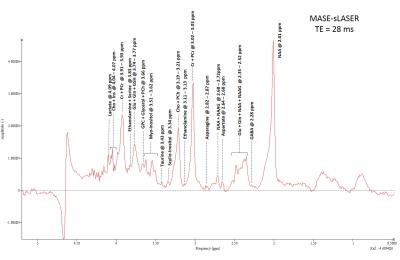
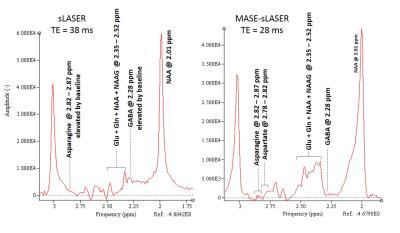
Figures 2 shows an example of high quality short TE=28 ms spectra acquired using MASE-sLASER from a voxel positioned at the occipital region of the brain. Major detected metabolites are labeled in this spectrum. Some low concentration metabolites including GABA, Asparagine and Ethanolamine are observable. For the sake of comparison, Figure 3 shows spectra obtained using MASE-sLASER (TE=28ms) and sLASER (TE=38ms) from the same voxel. Both spectra are zeroth order phase corrected to get an absorption lineshape for NAA. The range of frequencies between 2 -3 ppm are shown in the figure. The spectrum acquired with MASE-sLASER clearly has flatter baseline though acquired at 10 ms shorter TE, an observation for which there is no obvious explanation.Discussion
Shorter duration of the adiabatic refocusing SLR pulses of the MASE compared to conventional hyperbolic secant pulses with an acceptable bandwidth of about 3.5 KHz made it possible to achieve TE as short as 28 ms despite having one more RF pulse than the conventional sLASER sequence. The main feature of the MASE-sLASER compared to the conventional sLASER is its matched CSDE in all three directions. This is because of the totally matched slice selection gradients of the MASE pulses used for the single voxel selection in this sequence. The implemented short TE MASE-sLASER sequence has the same CSDE of 1.8 mm/ppm in all three directions. The other matched CSDE sequences generally available are full LASER and STEAM. Compared to MASE-sLASER, full LASER would lead to longer TE due to having one more RF pulse and STEAM lacks the advantage of full intensity signal. Moreover, we have observed some specific characteristics in the resultant spectra making MASE-sLASER sequence different than the conventional sLASER sequence. The resulting spectra with MASE-sLASER benefit from much lower baseline compared to the conventional sLASER sequence which is clear in figure 3 specifically between 2-2.3 ppm. Also, the pattern of the J coupled metabolites specifically appearing between 2-3 ppm is different between the two sequences. These two characteristics need further investigation.Conclusion
MASE-sLASER is a matched CSDE sequence with low CSDE providing more exact representation of the metabolites in the imaged voxel compared with the conventional sLASER which has different CSDE in one of the three slice selection directions. The very high quality of the acquired spectra with MASE-sLASER includes a flatter baseline compared with the conventional sLASER sequence.Acknowledgements
This work was funded by the Helmholtz Alliance ICEMED – Imaging and Curing Environmental Metabolic Diseases, through the Initiative and Networking Fund of the Helmholtz Association.References
1. Scheenen TW, et al. Short echo time 1H-MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses. Magn Reson Med. 2008;59(1):1-6.
2. Dyvorne H, et al. Ultrahigh field single-refocused diffusion weighted imaging using a matched-phase adiabatic spin echo(MASE). Magn Reson Med. 2016;75(5):1949-57
3. Gruetter R, Tkác I. Field mapping without reference scan using asymmetric echo-planar techniques. Magn Reson Med. 2000;43(2):319-23
4. Naressi A, et al. Java-based graphical user interface for the MRUI quantitation package. MAGMA. 2001;12(2-3):141-52.