2998
Pinpointing differences in diffusion characteristics of metabolites determined in human gray matter using simultaneous spectral and diffusion modeling1Depts. Radiology and Clinical Research, University Bern, Bern, Switzerland, CH-3010, Switzerland
Synopsis
A recently described non-water suppressed diffusion-weighting MR spectroscopy sequence was applied in 13 healthy volunteers. The method uses the large water signal to compensate motion-related signal drop for the metabolite signals. 2D signal modeling with FiTAID allows for implementation of different prior knowledge constraints in order to prevent non-physical solutions or to restrict the number of unknown diffusion constants. In gray matter, highly significant differences for ADCs of several metabolites (faster diffusion for glutamate than NAA or myo-inositol) were found and their dependence on prior knowledge constraints investigated.
Purpose
As shown recently1,
use of a non-water-suppressed (nWS) diffusion-weighted MR spectroscopy (DWS)
sequence allows correction for motion-induced signal reductions and thus improved
estimation of ADC values. Simultaneous 2D modeling of spectral features as well
as ADCs with FiTAID2 allows for determination of diffusion characteristics with
implementation of different prior knowledge constraints3, where
such constraints can be used to prevent non-physical solutions or restrict the
number of unknown diffusion constants. The purpose of this study was to determine
diffusion characteristics in human gray matter of as many metabolites as
possible and to investigate the influence of such prior knowledge constraints
using data from 13 subjects.Methods
A nWS DWS sequence based on metabolite-cycled STEAM with CSF nulling by water-selective inversion recovery1 was applied in 13 healthy volunteers in occipital gray matter (TI/TE/TM/TR= 1500/37/150/3500ms; diffusion gradient length δ=11ms, diffusion time Δ=168ms, maximum diffusion gradient Gmax=38 mT/m, maximal b-value bmax=5236 s/mm²) with water referencing to correct for motion effects. Metabolite ADC values were estimated in FiTAID2,3 based on mono-exponential diffusion. Estimated Cramer Rao lower bounds (CRLB) and variation of estimated ADCs over the cohort were used to judge the feasibility to determine ADCs of specific metabolites under specific prior knowledge restraints. The basis set comprised the 21 most abundant metabolites and a macromolecular baseline (MM).
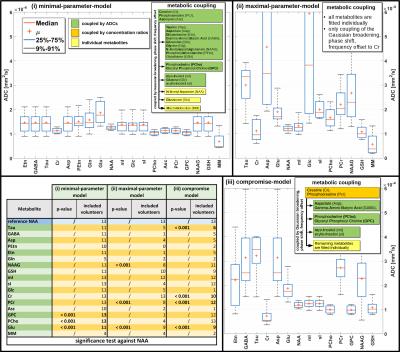
Modeling: besides usual spectral prior knowledge constraints, we investigated the benefit of linking ADCs for different metabolites. Three cases are presented: (i) minimal-parameter-model: only spectrally prominent metabolites or metabolite groups are bestowed with individual ADCs (NAA, Cr+PCr, cholines, sugars, glutamate, MM) while all others are forced into identical diffusion attenuation; (ii) maximal-parameter-model: individual ADC values are fitted for all metabolites; (iii) compromise-model, sought as a tradeoff between reduced fitting uncertainty and extended number of individual ADCs.
Statistics: ADC values were excluded from cohort average calculation if relative CRLB for area or ADC was >50%.
Results and Discussion
Fig. 1
shows the averaged DW spectra obtained from 13 subjects at different b-values.
Cohort fitting results for the three models are presented in Fig. 2. They
convey that glutamate clearly diffuses significantly faster than NAA (confirmed
by paired t-tests in case of all three models, as listed in Fig. 2). Table 1
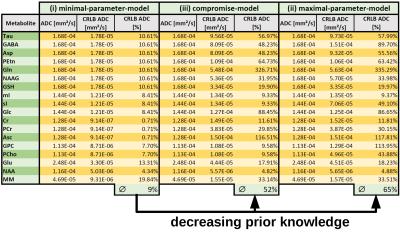
summarizes the fitting results of a single in vivo measurement with the
obtained CRLBs for model case (i) and those expected
for the case of released prior knowledge in models (ii) and (iii). Most
individual metabolite ADCs show a considerably increased uncertainty with lifted
prior knowledge. Still, it can be expected that individual ADCs of certain less
prominent metabolites can be reasonably well determined with the given data and
fitting models (e.g. glutamate, GSH or NAAG). This is mostly confirmed in the
cohort results (Fig. 2), where NAA, myo-inositol, glutamate and GSH show
fairly tight results even when equipped with individually fitted ADC values. Fig.
2 also lists the number of cases where interpretable results are obtained for
each metabolite (i.e. CRLB of area and ADC < 50%). In addition, it also
shows which metabolites are found to diffuse differently than NAA (paired
t-tests, listed only if n>5). Results depend on the fitting model. From
these findings, it appears that model iii) is a reasonable way to evaluate
these spectra with fairly tight ADC estimates for a number of metabolites and demonstration
of differences in diffusion constants between e.g. NAA and glutamate and
probably also taurine. In addition, from
Fig. 2 it does not seem that linking and equating the ADCs of the minor
metabolites would lead to substantial bias for the prominent metabolites, but
there is no substantial gain in robustness of estimation either. Interestingly,
linking merely the relative proportion of the areas (i.e. concentrations) for
strongly overlapping metabolites like Cr/PCr may result in very distinct
diffusion coefficients for these components (Cr/PCr in model (iii)). However, whether
or not the determined differences are real, remains questionable, since this is
almost equivalent to a biexponential fit of a single component.Conclusions
Diffusion
characteristics for metabolites in human gray matter were determined on a
clinical 3T system in a cohort of healthy subjects. ADCs for NAA, myo-inositol,
total cholines, total creatine, and MM, but also glutamate and GSH seem to be
well measurable in the cohort. Glutamate was found to diffuse significantly
faster than NAA, while other results may depend on the exact fitting model. It
was shown that linking ADCs for low concentration, multipattern metabolites may
stabilize the fit procedure, but care has to be used in interpretation of
results if any prior knowledge is used for combined modeling.Acknowledgements
This research was supported by the Swiss National Science Foundation (#320030_156952).References
1. Döring A, Adalid Lopez V, Brandejsky V, Boesch C, Kreis R, Diffusion weighted MR spectroscopy without water suppression allows to use water as inherent reference signal to correct for motion-related signal drop. In: Proc. Intl. Soc. Mag. Reson. Med. 24 (2016) #2395
2. Chong DGQ, Kreis R, Bolliger CS, Boesch C, Slotboom J. Two-dimensional linear-combination model fitting of magnetic resonance spectra to define the macromolecule baseline using FiTAID, a Fitting Tool for Arrays of Interrelated Datasets. Magn Reson Mater Phy 24:147-164 (2011).
3. Adalid Lopez VJ, Döring A, Kyathanahally SP, Bolliger CS, Kreis R. Simultaneous modeling of spectra and apparent diffusion coefficients. In: Proc. Intl. Soc. Mag. Reson. Med. 24 (2016) #2369.
Figures
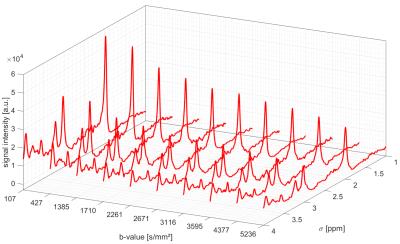
Fig. 1. Diffusion weighted spectra averaged over the whole cohort for 10 b-values, broadened with 2 Hz for visualization.