2991
Gannet 3.0: Developing post-processing tools for accelerated J-difference editing1Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2F. M. Kirby Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States
Synopsis
Initially conceived as
Purpose
To describe developments in the MATLAB-based processing package for edited magnetic resonance spectroscopy (MRS), Gannet(1). Originally conceived as a batch-processing tool for GABA-edited MRS data acquired with the MEGA-PRESS pulse sequence(2), Gannet has been extended to incorporate three new processing streams for measurements of glutathione (GSH)(3) and simultaneous editing of multiple metabolites and regions(4).Methods
In order to demonstrate the new functionalities, edited MRS experiments were acquired in two healthy volunteers. We have developed protocols for GSH editing(3), simultaneous editing of GABA in two brain regions with MEGA-PRIAM (MEGA parallel reconstruction in accelerated multivoxel MRS)(5), and simultaneous editing of GABA and GSH with HERMES (Hadamard encoding and reconstruction of MEGA-edited spectroscopy)(4). Experiments were performed on a Philips Achieva 3T with a 32-channel head-coil, as follows: GSH-edited MEGA-PRESS: TE 80 ms; editing pulses applied at 4.56 ppm (ON) and 8 ppm (OFF); GABA-edited MEGA-PRIAM: TE 68 ms; editing pulses applied at 1.9 ppm (ON) and 7.46 ppm (OFF); GABA/GSH-edited HERMES: TE 80 ms; editing pulses applied with a Hadamard encoding scheme at 4.56 ppm and 1.9 ppm for GSH and GABA, respectively. Additional parameters: 320 averages; 2048 datapoints; spectral width 2 kHz; 3x3x3 cm3 voxel (except 3.6x3.6x3.6 cm3 for HERMES); TR 2 s; editing pulse duration 20 ms (except 12 ms for MEGA-PRIAM); and VAPOR water suppression.
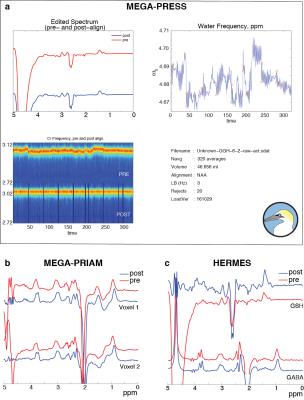
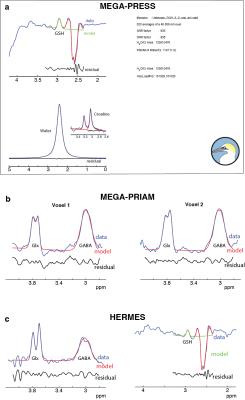
Frequency-and-phase correction (FPC) for GSH-editing: Frequency-and-phase correction (FPC) is a crucial step in the processing of J-difference-edited MRS(6). For GABA-edited MRS, spectral registration(7) is Gannet’s default method, correcting individual time-domain signals after modeling. Automated processing of GSH-edited MEGA-PRESS data requires a new FPC strategy because editing pulses partially suppress the water signal, biasing the registration of OFF to ON transients. Here, Gannet uses frequency-domain modeling of the NAA singlet to determine the frequency and phase shifts. The frequency-domain GSH difference spectrum is modeled between 2.35 and 3.3 ppm using a quadratic baseline, a Gaussian term to fit the GSH signal at 2.95 ppm and four Gaussians to model the co-edited aspartyl signals at ~2.6 ppm.
Reconstruction of two voxels: PRIAM uses SENSE reconstruction to separate the signals from two simultaneously dualband-excited voxels(8). PRIAM reconstruction has been implemented within Gannet, calculating the SENSE unfolding matrix from a B1-reference image in order to calculate separated time-domain data for each voxel. A ‘region’ level has been added to the Gannet data structure to accommodate multiple edited voxels per dataset. After PRIAM reconstruction, data proceeds through Gannet as usual; the default fitting routine for GABA-edited spectra now fits both GABA and Glx edited signals (2.79–4.10 ppm).
HERMES analysis: HERMES uses a Hadamard encoding editing strategy to run two (or more) MEGA-edited experiments simultaneously. Gannet performs the Hadamard reconstruction of separate edited spectra, and a ‘target’ level has been added to the Gannet data structure to accommodate multiple edited metabolite targets per dataset.
FPC for HERMES: FPC for HERMES spectra is especially challenging since there are four subspectra to co-register, compared to two with MEGA-PRESS. Additionally, the subspectra share fewer commonalities – editing impacts the water signal in GSH-ON scans and the NAA signal in GABA-ON scans, making most common strategies ineffective. Therefore, Gannet performs a hierarchical FPC: first, spectral registration is applied to all averages; second, the average GSH-ON, GSH-OFF, GABA-ON and GABA-OFF spectra are calculated; thirdly, the GSH-ON and GSH OFF spectra are aligned in the frequency domain to the GABA-OFF spectrum (range 2.8-3.53 ppm), and the GABA and GSH difference spectra are calculated. Additional developments include eddy-current correction(9) and SVD water filtering(10).
Results
Figure 1a shows the output of the GannetLoad module for GSH-edited MEGA-PRESS. Figure 1b and 1c show the top-left GannetLoad panel only for MEGA-PRIAM and HERMES data respectively. In each case, it can be seen that high-quality edited spectra are generated after FPC (blue spectra). Figure 2 shows the corresponding outputs from the GannetFit module, with the full output for GSH-edited MEGA-PRESS in a, and the top-left GannetFit panels only in b and c for MEGA-PRIAM and HERMES data.Discussion
With Release 3.0, Gannet expands its functionality to incorporate PRIAM and HERMES reconstruction, FPC, fitting for GSH-edited spectra, and an expanded data structure to accommodate multiplexed experiments. New editing approaches, particularly accelerated editing, demand new reconstruction and post-processing strategies. Here, we have demonstrated the implementation of appropriate pipelines in Gannet 3.0 to harness the potential of multiplexed editing.Conclusion
Gannet 3.0 provides new functionalities that permit post-processing of data from both conventional and accelerated editing techniques.Acknowledgements
This work was supported by NIH grants R01 EB EB016089, R01 MH106564 and P41 EB015909.References
1. Edden RA, Puts NA, Harris AD, Barker PB, Evans CJ. Gannet: A batch-processing tool for the quantitative analysis of gamma-aminobutyric acid–edited MR spectroscopy spectra. Journal of Magnetic Resonance Imaging 2014;40(6):1445-1452.
2. Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR in biomedicine 1998;11:266-272.
3. Terpstra M, Henry PG, Gruetter R. Measurement of reduced glutathione (GSH) in human brain using LCModel analysis of difference-edited spectra. Magnetic resonance in medicine 2003;50(1):19-23.
4. Saleh MG, Oeltzschner G, Chan KL, Puts NA, Mikkelsen M, Schär M, Harris AD, Edden RA. Simultaneous edited MRS of GABA and glutathione. NeuroImage 2016;142:576–582.
5. Oeltzschner G, Puts NA, Chan KL, Boer VO, Barker PB, Edden RA. Dual-Volume Excitation and Parallel Reconstruction for J-Difference-Edited MR Spectroscopy. Magnetic resonance in medicine 2016;DOI 10.1002/mrm.26536.
6. Evans CJ, Puts NA, Robson SE, Boy F, McGonigle DJ, Sumner P, Singh KD, Edden RA. Subtraction artifacts and frequency (Mis-) alignment in J-difference GABA editing. Journal of Magnetic Resonance Imaging 2013;38(4):970-975.
7. Near J, Edden R, Evans CJ, Paquin R, Harris A, Jezzard P. Frequency and phase drift correction of magnetic resonance spectroscopy data by spectral registration in the time domain. Magnetic resonance in medicine 2015;73(1):44-50.
8. Boer V, Klomp D, Laterra J, Barker PB. Parallel reconstruction in accelerated multivoxel MR spectroscopy. Magnetic resonance in medicine 2015;74(3):599-606.
9. Klose U. In vivo proton spectroscopy in presence of eddy currents. Magnetic resonance in medicine 1990;14(1):26-30.
10. Vanhamme L, Fierro RD, Van Huffel S, de Beer R. Fast removal of residual water in proton spectra. Journal of magnetic resonance 1998;132(2):197-203.
Figures