2941
Measuring lactate dehydrogenase activity with proton detected 13C hyperpolarization1Cancer Research UK Cambridge Institute, University of Cambridge, Cambridge, United Kingdom
Synopsis
Dissolution Dynamic Nuclear Polarized NMR (dDNP NMR) is a promising new tool for assessing metabolism in vivo. The signals of the hyperpolarized substrate and its downstream metabolites are usually detected by direct 13C observation. Here we demonstrate an effective way to repetitively transfer hyperpolarization via indirect couplings from [1-13C] to [3,3,3-1H3] in [1-13C] lactate formed from hyperpolarized [1-13C] pyruvate. The changes in the hyperpolarized [3,3,3-1H3] lactate peak were fitted to a kinetic model. The method sets the stage for dynamic hyperpolarized 1H imaging.
Purpose
There are advantages for 1H detection of 13C hyperpolarization including an approximately eight-fold increase in SNR compared to direct 13C detection [1]. Since direct hyperpolarization of 1H is difficult, mainly due to its fast T1 relaxation, the feasibility of transferring hyperpolarization from 13C to 1H nuclei and then acquiring the enhanced proton spectrum has been explored in vitro [2-4], in yeasts [5] and in rat brain [6]. Hyperpolarization transfer experiments have used either an spectrally unselective INEPT sequence, destroying all the 13C hyperpolarization in one shot, or have relied on cross relaxation, which yields a very low SNR. Here we acquired dynamic hyperpolarized 1H NMR spectra of [1-13C] lactate formed from [1-13C] pyruvate in the reaction catalyzed by lactate dehydrogenase (LDH) using repetitive spectrally selective INEPT [7] (ssINEPT). We were able to fit a kinetic model to the time course of the peak integral and could extract the apparent first order rate constant, kp, with the assumption that the [1-13C] pyruvate T1 relaxation time is known.Methods
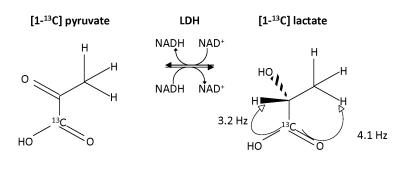
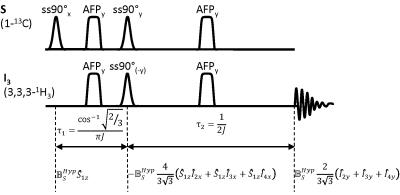
Because of the stronger coupling and the bigger chemical shift difference with water, polarization was transferred into the three magnetically equivalent C3 position protons in lactate (J13=4.1 Hz and σ=1.32 ppm) rather then the C2 proton (J12=3.2 Hz and σ=4.125 ppm) (Fig 1). The timing for an adapted INEPT sequence that transfers from one spin into three magnetically identical spins was determined (Fig 2). Using J13=4.1 Hz gives τ1=47.48 ms and τ2=121.95 ms. The spectrally selective 90° pulses were Gaussian shaped 7.1 ms pulses with a spectral width of 370 Hz (FWHM). The HS8 AFP pulses were 10 ms. To demonstrate that the proposed sequence is suitable for transferring polarization, experiments were performed with thermally polarized 37.5 mM [1-13C] lactate. To avoid signal originating from direct excitation of the observed nuclei, the ssINEPT sequence employed a form of phase cycling where in every other acquisition the phase of the initial 90° pulse as well as the readout phase of the receiver was shifted by 180°. Measurements were performed with TR=300 s and Navg=32. For the enzyme experiments, a buffer containing 37.5 mM NADH (pH 7.2, 120 mM Tris, 200 mM NaCL) was prepared. A stock enzyme solution was prepared containing 2000 U rabbit muscle LDH (Sigma Aldrich, Gillingham, UK), 1 ml glycerol and 1 ml buffer solution (pH 7.0) containing 20 mM Hepes, 200 mM KCl, 0.1 mM EDTA. From this stock solution 900 μl (=900 U) were added to 2 ml of the NADH containing buffer. Hyperpolarized [1-13C] pyruvate was prepared as described previously [8]. For a final concentration of 75 mM, the hyperpolarized [1-13C]pyruvate was dissolved in 6 ml buffer (pH 7.2, 40 mM Hepes, 94 mM NaOH, 30 mM NaCl and 100 mg/L EDTA). After starting the acquisition with TR =2 s, 2 ml of the pyruvate solution were injected into an NMR tube containing the enzyme solution in a 14 T vertical bore magnet. Phase cycling was not used in these experiments. The peak integral values SL(t) in each spectrum were determined using BRUKER topspin software. Starting with the values acquired after injection, the analytical solution of the two-site exchange saturation recovery model, neglecting backflow from lactate to pyruvate and lactate T1 relaxation is: $$ S_L(t)=z+A \exp{[-(k_p+1/T_{1P})t]} $$ Data were fitted to this equation. z, A and kP were left as fitted parameters. T1P was set fixed to 49.46 s, as described in the literature [9].Results
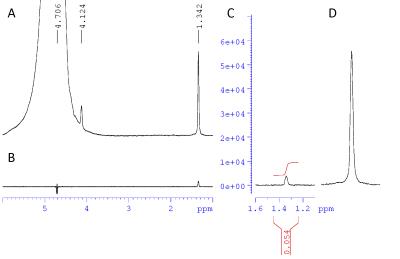
The peak proton integral following transfer of carbon polarization at thermal equilibrium differed by a factor of 0.054 from the proton integral obtained using a direct pulse and acquire sequence (Fig 3). Neglecting T2 decay the theoretically achievable value is $$$\frac{2}{3\sqrt{3}}\frac{\gamma_{13C}}{\gamma_{1H}} \approx 0.097$$$.
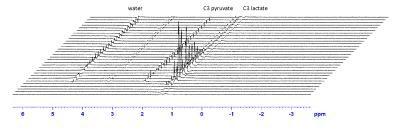
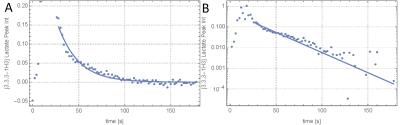
We therefore estimate the polarization transfer to work at 56% efficiency. The time course of the ssINEPT hyperpolarized acquisitions (Fig. 4) showed the expected exponential decay (Fig. 5). Fitting the kinetic model to the dataset yielded a rate constant, kp= (0.024±0.002) s-1 .
Discussion
The losses in the thermal polarization transfer experiments can be explained by T2 decay for both 1H and 13C during the pulse sequence. An efficiency of 56% would still give a four fold increased SNR compared to direct 13C detection. The enzyme experiment showed the expected time course behaviour and could be fitted very well by the model.Conclusion
Proton detected dDNP has the potential to be used to assess enzymatic activity. The technique could be adapted for in vivo MR imaging.Acknowledgements
The work was supported by a Marie Curie Action ITN (EUROPOL) and a Cancer Research UK Programme grant.References
1. Levitt M. Spin Dynamics: Basics of Nuclear Magnetic Resonance. Wiley; 2001.
2. Harris T, Giraudeau P, Frydman L. Kinetics from indirectly detected hyperpolarized NMR spectroscopy by using spatially selective coherence transfers. Chem - A Eur J. 2011;17(2):697-703.
3. Sarkar R, Comment A, Vasos P, et al. Proton NMR of 15N-Choline Metabolites Enhanced by Dynamic Nuclear Polarization. J Am Chem Soc. 2009;131(44):16014-16015.
4. Chekmenev E, Norton V, Weitekamp D, Bhattacharya P. Hyperpolarized 1H NMR employing low γ nucleus for spin polarization storage. J Am Chem Soc. 2009;131(9):3164-3165.
5. Dzien P, Fages A, Jona G, et al. Following Metabolism in Living Microorganisms by Hyperpolarized 1H NMR. J Am Chem Soc. 2016;138(37):12278-12286.
6. Mishkovsky M, Cheng T, Comment A, et al. Localized in vivo hyperpolarization transfer sequences. Magn Reson Med. 2012;68(2):349-352.
7. Morris G, Freeman R. Enhancement of Nuclear Magnetic Resonance Signals by Polarization Transfer. J Am Chem Soc. 1979;233(2):760-762.
8. Day S, Kettunen M, Gallagher F, et al. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat Med. 2007;13(11):1382-1387.
9. Daniels C, Mclean M, Schulte R, et al. A comparison of quantitative methods for clinical imaging with hyperpolarized 13C-pyruvate. NMR Biomed. 2016;29(2):387-399.
Figures