Leo L. Cheng1, Emily A. Decelle2, Johannes L Kurth2, Shulin Wu3, Taylor L. Fuss2, Lindsey A. Vandergrift2, Elita M. DeFeo2, Elkan F. Halpern2, Matthias L Taupitz2, W. Scott McDougal2, Aria F. Olumi2, and Chin-Lee Wu2
1Molecular Pathology, Massachusetts General Hospital, Charlestown, MA, United States, 2Massachusetts General Hospital, Charlestown, MA, United States, 3Massachusetts General Hospital, Boston, MA, United States
Synopsis
While
serum prostate specific antigen (PSA) testing improved early detection of
prostate tumors, implementation of this tool also created a large patient population
in which identified cancer lesions were actually indolent. For patients who
choose to undergo prostatectomy, cancer aggressiveness can only be determined
by post-procedure pathology analysis of cancerous tissue. Additionally, cancer
recurrence predictions are often unreliable. As an alternative method to aid
decisions regarding treatment, we sought to develop MRS tools which can predict
cancer aggressiveness based on the novel use of histologically benign (Hb)
tissue.
Introduction
The
clinical challenge of prostate cancer (PCa) arises due to the fact that over
70% of patients diagnosed by the gold standard of serum prostate specific
antigen (PSA) screening will have an indolent form of cancer with little effect
on their well-being. More aggressive cancer which does harm well-being is found
in about 12% of PSA-diagnosed patients. For the patients in this group who do
undergo prostatectomy, tumor grade can only be determined post-operatively. Furthermore,
current predictive nomograms are fraught with uncertainty about likelihood of
cancer recurrence. Under the hypothesis that metabolomic fields within
cancerous tissue can cause metabolite delocalization into neighboring benign
tissue, we aimed to develop biomarkers to distinguish indolent from aggressive PCa
using histologically benign (Hb)
tissues of PCa patients. Implementation of such a diagnostic could prevent
overtreatment and create more individualized therapy plans. Methods
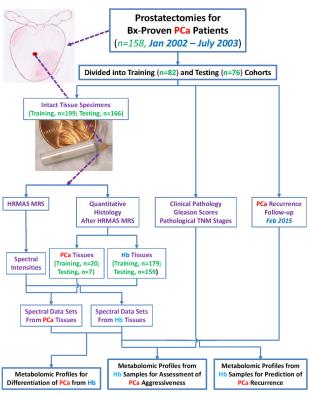
After
radical prostatectomy, 365 tissue samples were collected from 158 cancerous
prostates. They were snap-frozen in liquid nitrogen <45 minutes after
radical prostatectomy and stored at -80oC until MRS analyses. Overall
experimental design for characterization of tissue is diagrammed in Figure 1. Intact tissue MRS. Tissue scans were conducted with high-resolution magic angle spinning on a Bruker
AVANCE 600MHz spectrometer. Samples were weighted (~10mg) and placed into a 4mm
rotor with plastic inserts to create a 10µl sample space; 1.0µl D2O
was added for field locking. Spectra were recorded at 4ºC with a repetition
time of 5s, and a rotor-synchronized DANTE protocol conducted with spinning at
both 600 and 700Hz. Data were analyzed using AcornNMR-Nuts with relative
intensities obtained from integrated intensities normalized by the spectral
intensity between 0.5 and 4.5ppm. Quantitative
histopathology. Tissue was processed and analyzed by traditional
histopathology. Quantification of volume percentages of pathological features
and PCa/Hb categorization of tissue was performed by a pathologist. Results
Analyses
indicate the ability of MRS to determine cancer aggressiveness. The 3.60ppm spectral
region in Hb prostatectomy tissue can differentiate between PCa prognostic
grade group (PGG)1&2 and PGG3&4 groups, with an overall accuracy of 73%
and 71% for the training and testing cohorts. Tumor groups, specifically
pT=IIab and pT=IIac, can also be distinguished using the lipid spectral region
of 0.93-0.96ppm. Further analysis showed that the 3.60ppm region and principal
component (PC) 4 could distinguish a subgroup of low malignant potential from
the main group. Application
of these PC4 parameters to a testing cohort showed that 10 of 58 (17%) of low
aggressive cases could be distinguished among all 72 (14%) cases. Lastly, MRS
measurements of Hb tissue help predict tumour biochemical recurrence (BCR).
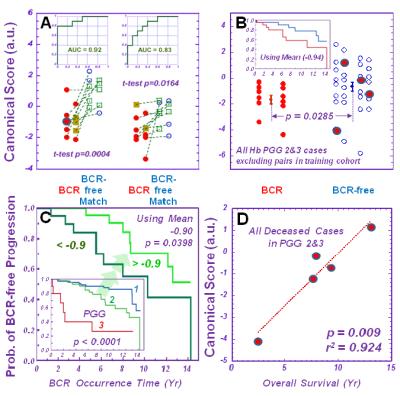
Principal components and canonical analyses calculated from 36 spectral regions
of ten pairs of BCR and non-BCR cases with the training cohort can differentiate
BCR from non-BCR cases within the testing cohort with an accuracy of 83% (Figure 2A). Given that the canonical
score was comprised primarily of PGG 2 and 3 cases, applying the canonical
score calculations onto the PGG 2 and 3 cases resulted in a statistically
significant differentiation between BCR and non-BCR cases (Figure 2B). Within the PGG 2 group only, the BCR discriminant
canonical values can further differentiate the group into two (Figure 2C) according to Kaplan-Meier
analysis. The canonical score also linearly correlate with patient survival
time (r2 = 0.924) (Figure 2D). Discussion and Conclusions
Where
pathological analysis of a benign core might present a false negative due to
the heterogeneity of PCa, the ability of the spectral region to distinguish
between cancer grades could enable MRS to detect of aggressive forms of cancer
when pathology fails to do so. Currently tumor grade can only be determined
after radical prostatectomy, but the capability of the spectral regions in Hb
tissue to determine tumor aggressiveness could inform treatment decisions prior
to surgical intervention. Even with imperfect characterizations, identification
of a distinct low malignancy potential group could better inform the therapy
plan for 110,000 patients annually worldwide. Nomograms today remain incapable
of differentiating between matched cases, so the ability of canonical scores
from our matched BCR and non-BCR cases to predict likelihood of cancer
recurrence and survival time highlights their potential as a clinical tool. These
findings demonstrate the ability of MRS-measured metabolomic fields in Hb tissue to detect biochemical changes,
and, as a result, determine disease aggressiveness. We present their potential
utility as diagnostic tools to better inform treatment decisions in the PCa
clinic.Acknowledgements
Authors
acknowledge support by PHS NIH grants CA115746, CA115746S2, and
CA162959 and the A. A. Martinos Center for Biomedical
Imaging.References
No reference found.