2871
Feature-Tracking for Volume and Strain with Subtly Tagged SSFP1Physiology & Experimental Medicine, University of Toronto, Toronto, ON, Canada, 2Siemens Healthcare, Erlangen, Germany, 3Anatomy and Medical Imaging, University of Auckland, Auckland, New Zealand
Synopsis
A novel subtly tagged cardiac acquisition coupled with non-rigid registration is developed and tested in healthy volunteers. The technique produces myocardial strain characterization and ventricular volumetrics within a single scan.
Purpose:
While myocardial strain measurements from MRI tagging have been shown to be a useful biomarker in a number of clinical applications1, tagging creates one- or two-dimensional saturation bands in the image and thus makes them unsuitable for ventricular functional analysis. To evaluate both cardiac ventricular function (e.g. volumes and mass) and heart wall strain, two separate scans must traditionally be performed: cine tagged gradient-recalled echo (GRE) and cine untagged steady-state free precession (SSFP). In clinical practice this increases the number of breath-holds and thereby on-the-table scan time for cardiac patients, many of whom have difficulty with repeated breath holds.
More recently, non-rigid deformable registration methods have been applied to untagged SSFP images to calculate strain2-4. This technique tracks texture patterns from frame to frame, producing deformation fields that enable the calculation of myocardial strain. Performed without an additional tagged GRE sequence, untagged SSFP strain reduces total scan acquisition time while providing ventricular functional parameters. Although global strain averaged over a slice agrees well with tagged GRE, segmental strains show statistically significant discrepancies due to lack of features in the myocardium, with overestimation in the septal wall and underestimation in the lateral wall5. As ischemic cardiac disease is a regional phenomenon, accurate localized results are required clinically.
Other works have investigated using tagged GRE images to evaluate global cardiac function by employing additional k-space and image space processing6. These results showed good LV mass correlation between untagged SSFP and processed images, yet image quality suffered in GRE compared with SSFP.
Here we aim to investigate whether the introduction of faint myocardial features (“subtly tagged SSFP”), paired with non-rigid deformable registration methods, can improve accuracy of regional myocardial strain and maintain accuracy of ventricular function.
Methods:
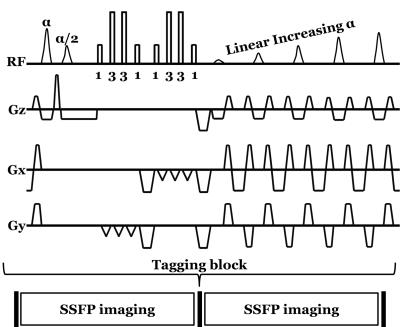
Imaging acquisition: The subtly tagged SSFP acquisition employs a prototype version of LISA-SSFP7 tagging. The tagging block (Figure 1) is composed of an α/2 flipback pulse at the detection of the ECG trigger, the implementation of a 1-3-3-1 binomial pulse structure in two orthogonal directions to produce grid tagging, gradient spoiling, and 5 linearly increasing start-up angles before the SSFP imaging block is resumed. The sequence further supports retrospective cardiac gating and parallel imaging (iPAT).
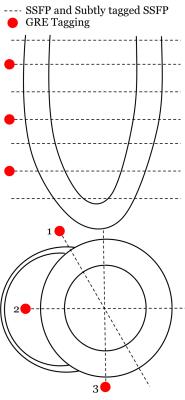
Whole-heart assessment – comparison to clinical scans: Fifteen healthy volunteers (42 ± 18 years old, 9 female) were imaged following informed consent using a clinical 3T MRI (Magnetom Skyra, Siemens Healthcare, Germany). Standard untagged SSFP imaging was performed in 7 short-axis slices and 3 long-axis views with α = 50⁰ (Figure 2). In these same slice locations, subtly tagged SSFP was performed, here choosing α = 40⁰ and total tagging angle = 46⁰. These parameters were chosen based on preliminary studies of tagging persistence to end-systole (ES)8, myocardial – blood pool contrast (to match standard untagged SSFP), and relative tag subtlety to the naked eye in unenhanced images. To as closely match the two imaging strategies as possible, both acquisitions had the same timing and spatial imaging parameters. Additionally, tagged GRE was acquired at 3 of the 7 short-axis slices equally spaced to produce basal, mid, and apical tagging series, and in the long-axis views.
Assessment of volume and strain: Reconstructed image sets for all volunteers were assessed by a single analyst using the cardiac image modeller tool (CIM, University of Auckland, New Zealand). After manual delineation of the end-diastolic (ED) endocardium and epicardium, images were automatically tracked through time using non-rigid deformable registration methods. For volume assessment, a 3D whole ventricle mesh was produced and tracked through time3.
Strain was estimated in a pixel-wise manner to produce time-resolved regional strain estimates. This was performed in the mid-ventricular short-axis slice from tagged GRE, and the corresponding untagged SSFP and subtly tagged SSFP slices. ES circumferential strain was assessed: averaged over the entire mid-ventricular slice, and in the 6 AHA segments according to anterior and posterior insertions of the right ventricle in the septum.
Statistics: For strain measurements, tagged GRE was used as the ‘gold standard’, and differences across regions and methods of measurement were compared using paired t-tests. Similarly, left ventricular volume measures from untagged SSFP and subtly tagged SSFP were compared using paired t-tests. Multiple comparisons (in regions for strain or across volume measures) were corrected using false discovery rate9, and statistical significance was established at the 5% level (p < 0.05).
Results:
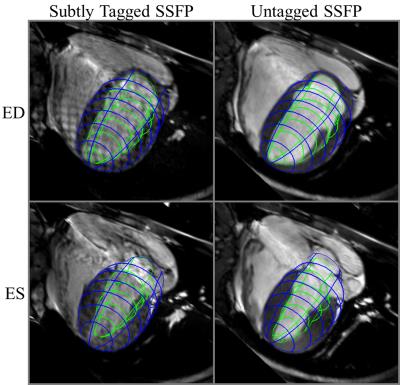
Compared with untagged SSFP (Figure 3), the myocardium – blood pool contrast in subtly tagged SSFP allowed for very similar endo- and epicardial contours to be traced and tracked through time, all while maintaining a grid pattern into ES.
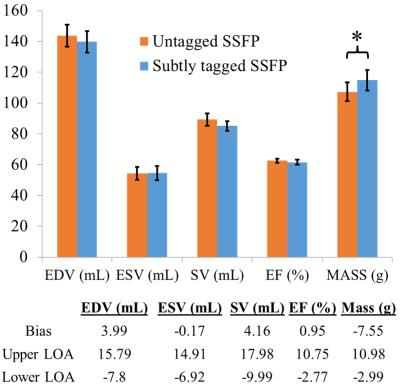
Figure 4 displays left ventricular volume analysis between untagged SSFP and subtly tagged SSFP. No significant differences were found for end-diastolic volume, end-systolic volume, stroke volume, and ejection fraction. ED mass was found to be statistically significant, with a slight overestimation using subtle tagging.
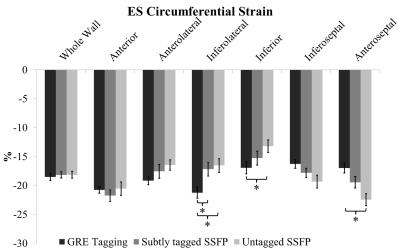
Figure 5 displays ES circumferential whole wall and regional strains across all acquisition types. Subtly tagged SSFP appears to outperform untagged SSFP. Significant differences were only observed in the inferolateral region, whereas untagged SSFP showed statistically significant overestimation in the septal wall and underestimation in the lateral wall.
Conclusion:
Subtly tagged SSFP enables more accurate regional strain assessment than untagged SSFP while maintaining accuracy of ventricular function in a single MRI scan. Previously either time (two scans) or accuracy of strain (as in the case of untagged SSFP strain measurements) had to be sacrificed in the assessment of both quantities. Future work will employ interobserver testing, as well as apical, basal, and longitudinal strain comparisons with tagged GRE.
While subtly tagged SSFP provided improved strain estimation, regional differences with GRE tagging were not totally resolved. Fine tuning could provide an optimized sequence for image quality, and strain and volume assessment.
Acknowledgements
We gratefully acknowledge Siemens Healthcare for their research support.References
1. Ibrahim el SH. Myocardial tagging by cardiovascular magnetic resonance: evolution of techniques--pulse sequences, analysis algorithms, and applications. J Cardiovasc Magn Reson 2011; 13: 36.
2. Augustine D, Lewandowski AJ, Lazdam M, Rai A, Francis J, Myerson S et al. Global and regional left ventricular myocardial deformation measures by magnetic resonance feature tracking in healthy volunteers: comparison with tagging and relevance of gender. J Cardiovasc Magn Reson 2013; 15: 8. 3. Li B, Young AA, Cowan BR. GPU accelerated non-rigid registration for the evaluation of cardiac function. Med Image Comput Comput Assist Interv 2008; 11(Pt 2): 880-7.
4. Wu L, Germans T, Guclu A, Heymans MW, Allaart CP, van Rossum AC. Feature tracking compared with tissue tagging measurements of segmental strain by cardiovascular magnetic resonance. J Cardiovasc Magn Reson 2014; 16: 10.
5. Cowan BR, Peereboom SM, Greiser A, Guehring J, Young AA. Image Feature Determinants of Global and Segmental Circumferential Ventricular Strain From Cine CMR. JACC Cardiovasc Imaging 2015; 8(12): 1465-6.
6. Makram AW, Khalifa AM, El-Rewaidy H, Fahmy AS, Ibrahim el SH. Assessment of cardiac mass from tagged magnetic resonance images. Jpn J Radiol 2016; 34(2): 158-65. 7. Zwanenburg JJ, Kuijer JP, Marcus JT, Heethaar RM. Steady-state free precession with myocardial tagging: CSPAMM in a single breathhold. Magn Reson Med 2003; 49(4): 722-30.
8. Markl M, Scherer S, Frydrychowicz A, Burger D, Geibel A, Hennig J. Balanced left ventricular myocardial SSFP-tagging at 1.5T and 3T. Magn Reson Med 2008; 60(3): 631-9.
9. Glickman ME, Rao SR, Schultz MR. False discovery rate control is a recommended alternative to Bonferroni-type adjustments in health studies. J Clin Epidemiol 2014; 67(8): 850-7.
Figures