2866
Three-dimensional Bi-ventricular Myocardial Feature Tracking for Congenital Heart Disease Using Standard Cardiac Cine MRI with Interpolation Technique Based on Moving Gradients1Department of Radiological Technology, Faculty of Fukuoka Medical Technology, Teikyo University, Omuta, Japan, 2Department of Clinical Radiology, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan, 3Division of Radiology, Department of Medical Technology, Kyushu University Hospital, Fukuoka, Japan, 4Department of Health Sciences, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan, 5Department of Diagnostic Imaging & Nuclear Medicine, Tokyo Women’s Medical University, Tokyo, Japan
Synopsis
Patients with congenital heart disease (CHD) often have complicated ventricular motion and configuration. Myocardial feature tracking (MFT) MRI can quantitatively analyze 2-dimensional myocardial motion. Although three-dimensional (3D) MFT is required for the patients with CHD, standard cine MRI is limited the setting of thin slice thickness attributed to increasing scan time. In this study, we analyzed bi-ventricular function with 3D MFT using only standard cine MRI datasets reconstructed by moving gradients based image interpolation technique. As the result, 3D MFT is useful to evaluate bi-ventricular function for patients with CHD, and can be easily applied to routine clinical MR examination.
Purpose
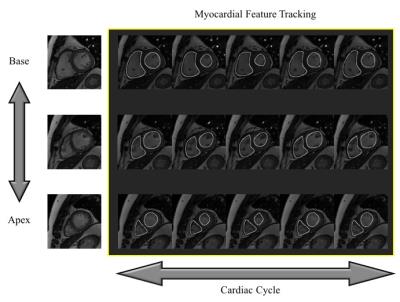
One of the great advantage of cardiac cine magnetic resonance imaging (MRI) to echocardiography is to provide as excellent visualization of bi-ventricular motion and structures with a wide field of view. Therefore, cardiac cine MRI has been generally used in clinical studies and demonstrated its accuracy and reproducibility for evaluation of left ventricular (LV) and right ventricular (RV) functions.1 As shown in Figure 1, myocardial feature tracking (MFT) MRI can quantitatively analyze 2-dimensional myocardial motion in any slice orientations and cardiac regions.2 In the patients with congenital heart disease (CHD), complicated ventricular motion and configuration are presented in a lot of cases. Although three-dimensional (3D) MFT is required for the patients with CHD, standard cine MRI is limited the setting of thin slice thickness such as computed tomography attributed to increasing scan time. Accordingly, approximately 10 stacked cine images are obtained with short-axis orientation in clinical MR examination. Previously, a path based method image interpolation technique with moving gradients was developed.3 This is the method for plausible interpolation of images, with a wide range of applications like temporal up-sampling for smooth playback of lower frame rate video, smooth view interpolation, and animation of still images. We hypothesized that standard stacked cine MRI can be interpolated as volumetric image data with this technique. In this study, we analyzed LV and RV function with 3D MFT using only standard cine MRI datasets reconstructed by moving gradients based image interpolation technique.
Methods
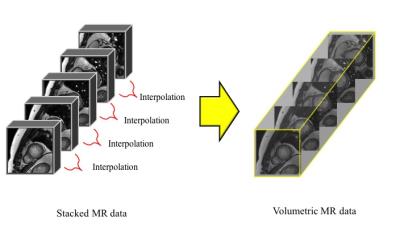
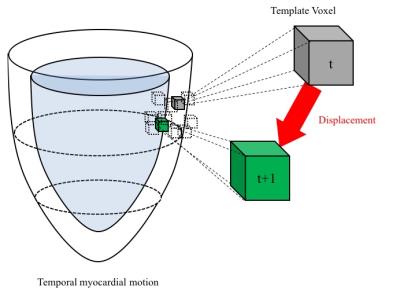
Datasets of short-axis cine MR images in 33 subjects (12 ± 4 years-old) including the patients with CHD from York University were analyzed.4 All cine images were obtained by steady state free precessions sequence. Spacing between slices per slice were set 7.0 to 13.0 mm. These stacked MR data were interpolated as approximately 100 volumetric MR images with moving gradients based technique for a cardiac cycle (Fig. 2). In the interpolated MRI, LV and RV myocardial borders were manually delineated by an experienced cardiac radiologist. The 3D displacement of each voxel of myocardial borders were calculated as the scalar quantity of a vector of the distance (mm) of the shifted voxel per a frame with the template matching technique (Fig. 3). Ventricular global displacement (GD) was defined the mean of the displacement of each voxel. The maximum GD of LV and RV for a cardiac cycle were compared to ejection fraction (EF) of LV and RV by Pearson correlation coefficients in all 33 subjects.Results
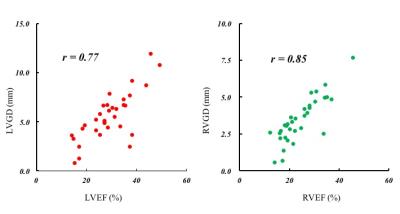
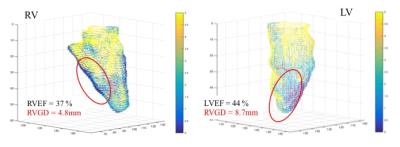
The mean ± SD of GD and EF for LV were 29.2 ± 8.8 % and 5.5 ± 2.4 mm, and those for RV were 24.4 ± 7.6 % and 3.4 ± 1.5 mm, retrospectively. Significant positive correlations were observed in all subjects between the LVGD and LVEF (r = 0.77, p <0.01) and between the RVGD and RVEF (r = 0.85, p <0.01) (Fig. 4). The representation of the 3D voxel plot of LV and RV myocardial border was shown in Figure 5 (LVGD = 8.7 mm vs. LVEF = 44 %, RVGD = 4.8 mm vs. RVEF = 37 %).Discussion
The GD of LV and RV were significantly correlated with LVEF and RVEF, respectively. As shown in Figure 5, 3D MFT could be clearly visualized the bi-ventricular motion and structure. Moreover, 3D MFT could detect the dysfunction area simply even if EF was preserved. Therefore, 3D MFT is useful diagnostic tool for the evaluation of bi-ventricular function. In this study, 3D myocardial function of LV and RV in patients with CHD could be analyzed with only standard cine MR images. This analysis would be able to perform repeatedly for CHD patients without radiation exposure such as follow up in growth process. Moreover, this analysis could be easily applied to routine clinical MR examination without special MR systems and sequences. Therefore, 3D MFT enables to analyze the bi-ventricular complex motion and provides the further information of dynamics for patients with CHD.Conclusion
In conclusion, 3D MFT is useful to evaluate the bi-ventricular function for the patients with CHD, and can be easily applied to routine clinical MR examination.Acknowledgements
This work was supported by JPS KAKENHI Grant Number JP16K19860.References
1. Sechtem U, Pflugfelder PW, Gould RG, et al. Measurement of right and left ventricular volumes in healthy individuals with cine MR imaging. Radiology 1987;163(3): 697–702.
2. Kawakubo M, Nagao M, Kumazawa S, et al. Evaluation of ventricular dysfunction using semi-automatic longitudinal strain analysis of four-chamber cine MR imaging. Int J Cardiovasc Imaging 2016; 32(2): 283-9.
3. Mahajan D, Huang F, Matusik W, et al. Moving gradients: a path-based method for plausible image interpolation. ACM computer graphics, SIGGRAPH 2009 In Annual conference series.
4. Andreopoulos A, Tsotsos JK. Efficient and Generalizable Statistical Models of Shape and Appearance for Analysis of Cardiac MRI. Med Image Anal 200; 12(3): 335-57.
Figures