2566
See It, Slice It, Learn It: Combined Ultra High Field MRI and High-resolution CT for an Open Source Virtual Anatomy Resource1Melbourne Brain Centre Imaging Unit, Department of Anatomy and Neuroscience, University of Melbourne, Parkville, Australia, 2Department of Anatomy and Neuroscience, University of Melbourne, Parkville, Australia, 3Harry Brookes Allen Museum, Department of Anatomy and Neuroscience, University of Melbourne, Parkville, Australia, 4Learning Environments, University of Melbourne, Parkville, Australia, 5Siemens Healthcare, Melbourne, Australia
Synopsis
Grasping human head and neck anatomy can be challenging for students, scientists and health professionals in medical disciplines. Ultra-high field, 7T MRI can create high-resolution images with multiple contrasts, revealing structural detail not easily apparent in dissection specimens. While widely exploited in clinical and scientific studies, its use has been limited so far in creating tools for medical education. We present a multi-modal combination of ex-vivo MRI and CT to create a high-quality head and neck anatomy resource to enhance cadaveric cross-sectional anatomy teaching.
Purpose and Introduction
Visualising 3D human head and neck anatomy can be challenging for students in medical disciplines. Ultra-high field (UHF), 7T MRI can create high-resolution images with multiple contrasts, revealing structural detail not readily apparent in dissection specimens. While ex-vivo UHF MRI has been readily exploited in clinical and scientific studies (1,2), its use has been more limited so far in creating interactive tools for anatomical education, focusing on static atlases (3,4). Additionally, while greater emphasis is now placed on interpretation of radiological anatomy in many curricula, detailed images from high-field systems are not widely available to medical educators. Moreover, the full 3D datasets generated by MRI are amenable to digital segmentation and annotation of their anatomical structures for identification, physical 3D printing and integration into emerging virtual-reality environments, for headset and interactive display. This is of growing interest for anatomy education (5,6). We present a high-technology resource for medical education that combines state of the art imaging, 3D modelling, VR and physical anatomy expertise, to produce an interactive teaching resource for head and neck anatomy. This work may help support many anatomy students at our institution and beyond.Methods
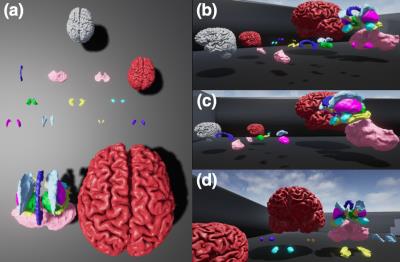
Specimen preparation: An unfixed head and neck specimen of an 86 year-old male donor with no gross brain pathology (approximate post-mortem interval of 10 days prior to first MRI) was used in this work. All cadaveric imaging was approved by the University of Melbourne Human Research Ethics Committee. The specimen was carefully maintained at 4°C immediately post-mortem, and between imaging sessions. Imaging: MRI was performed on a research 7 Tesla MRI system (Siemens Healthcare, Erlangen, Germany), and 400µm isotropic (the highest possible resolution with single-volume-excitation, whole-brain coverage on our system) anatomical MRI sequences were acquired including: T1-w (3D MP2RAGE: TE/TR/TIs/FA/NSA:2.4ms/4.9s/0.7+2.7s/5+6°/4avgs, and T2-w (3D SPACE: TE/TR/ETL/NSA:153ms/3s/140/1,GRAPPA 3+2). Due to these technical limitations on resolution, we also explored obtaining higher resolution at a second imaging session 2 weeks later, acquiring additional slab-selective T2-w images at 290µm isotropic (as above with TE 215ms, NSA 3) for anatomical comparison and SNR assessment (3 slabs for whole-brain coverage). CT was performed on a 128-slice Biograph mCT system (Siemens Healthcare, Erlangen, Germany), during session 1, with thin-slice image volumes acquired at 3 tube voltages (80kV, 120kV, 140kV, resolution:520x520µm, 0.1mm slice thickness). Image Processing: Linear image registration of datasets into a common space was performed using niftyReg (TIG UCL, London). The T1 MP2RAGE anatomical volume was then processed automatically using Freesurfer (v.6 development build July 2016, MGH) to produce 3D brain segmentations. These were then imported into Blender (Blender Institute, Amsterdam) for manual correction and visualisation and exported into Unreal Engine (v4. Epic Inc.) environment for final display.Results
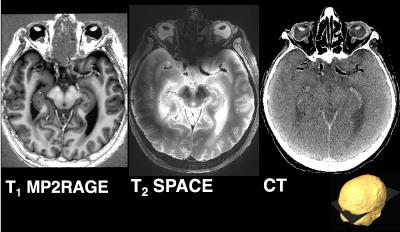
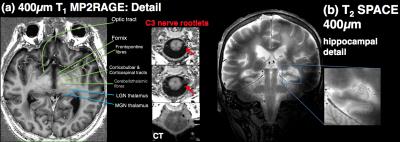
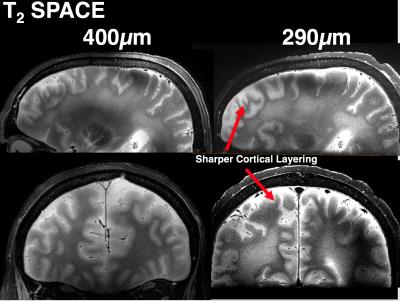
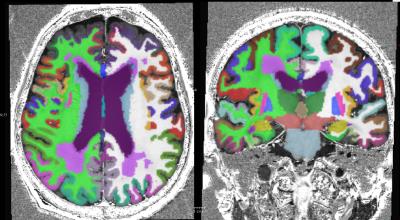
As shown in figure 1, T1 and T2-weighted MRI volumes were amenable to co-registration to CT data with good agreement of structures across modalities. 400µm T1 and T2-weighted images acquired at session 1 allowed identification of fine anatomical structures not readily seen on in-vivo imaging, including: individual cortical layers, hippocampal sub-regions and individual cervical spinal cord nerve rootlets. Comparing 400µm isotropic and 290µm isotropic slab-selective images from session 2 (a ~26 fold smaller voxel volume) allowed greater visualisation of fine structure, particularly cortical layers (appreciated on T2 SPACE data shown in figure 3). SNR reduced in grey matter from ~202:1 to ~85:1 and WM ~142:1 vs. 58:1. Automated Freesurfer segmentation produced plausible structural volumes with some misidentification of temporal and frontal regions (Figure 4). Segmented volumes were amenable to editing for minor corrections in Blender prior to import into Unreal Engine (Figure 5).Discussion and Conclusion
We have demonstrated that ex-vivo 7T MRI at 400µm was able to offer impressive anatomical detail and soft tissue contrast in a variety of brain and CNS structures with very high SNR. This enabled delineation of very fine anatomical structures down to fine spinal cord nerve roots. Whole-brain 400µm T1 Freesurfer segmentations provided a good initial starting point for segmentation of brain structures but required manual correctionof frontal and temporal regions to create satisfactory models. This was then readily exported into further open-source 3D software for editing and VR placement. The level of detail in these images should enable detailed further segmentation. Expanding this work, we also intend to incorporate diffusion tractography images into our atlas space and the VR environment.Acknowledgements
We are grateful to: University of Melbourne Learning and Teaching Initiative for project grant support; JOC is supported by a University of Melbourne McKenzie Fellowship; Siemens Healthcare for technical assistance and access to prototype MRI sequences. The MBCIU 7T MRI and PET/CT systems are supported by the Australian National Imaging Facility (NIF). High-performance computing support by the Multi-modal Australian ScienceS Imaging and Visualisation Environment (MASSIVE).References
1) Foxley S et al. Improving diffusion-weighted imaging of post-mortem human brains: SSFP at 7T Neuroimage. 2014 Nov 15; 102: 579–589.
2) Simons D et al. 7T MRI based innovative bleeding screening in forensic pathology: acquisition of traumatic (micro) bleeding in the brain. ECR Congress 2015 C-0385
3) Cho ZH (ed.) 7.0 Tesla MRI Brain Atlas: In Vivo Atlas with Cryomacrotome Correlation. Springer 2015,
4) Fatterpekar GM et. al MR microscopy of normal human brain. Magn Reson Imaging Clin N Am. 2003 Nov;11(4):641-53.
5) de Faria JW, et al. Virtual and stereoscopic anatomy: when virtual reality meets medical education. J Neurosurg. 2016 Nov;125(5):1105-1111
6) Baskaran V, et al. Applications and Future Perspectives of the Use of 3D Printing in Anatomical Training and Neurosurgery. Front Neuroanat. 2016 Jun 24;10:69
Figures