2557
Histological Validation of Diffusion Basis Spectrum Imaging Using Autopsied Multiple Sclerosis Brain Specimens1Department of Radiology, Washington University, St. Louis, Saint Louis, MO, United States, 2Department of Radiology, Washington University, St. Louis, 3Department of Neurology, Washington University, St. Louis, MO, United States, 4Advanced Imaging Research Center, Oregon Health & Science University, OR, United States, 5Radiological Health Sciences, School of Health Sciences, Purdue University, West Lafayette, IN, United States, 6Department of Radiology, Washington University, St. Louis, MO, United States
Synopsis
In this study, we demonstrate diffusion basis spectrum imaging (DBSI) is able to detect, differentiate and quantify different coexisting pathologies, particularly axonal loss and demyelination within autopsied multiple sclerosis human brain specimens. We correlated the DBSI derived maps with quantitative histology maps generated by means of color based segmentation. DBSI-derived fiber fraction was seen to correlate with Bielschowsky’s silver stain for axonal integrity, and the DBSI-derived radial diffusivity negatively correlated with Luxol Fast Blue-Periodic Acid-Schiff (LFB-PAS) stain for myelin integrity.
Introduction
The primary pathologies that underlie multiple sclerosis (MS) are axonal injury/loss, demyelination and inflammation. The roles that each of these individual pathologies play in disease progression are ill-defined. To validate MRI-derived white matter pathological metrics, it is necessary to quantify corresponding histological staining of cross-sectional sections of white matter tracts by counting of axons, myelin sheaths, or nuclei. A previous study1, for quantification of MS related changes within the autopsied spinal cord specimens revealed that diffusion basis spectrum imaging (DBSI)-derived radial diffusivity negatively correlated with Luxol Fast Blue-Periodic Acid-Schiff (LFB-PAS) positive staining, while DBSI-derived fiber fraction was seen to correlate with Bielschowsky’s silver positive staining. However this method, relied on manually counting positive staining within the down sampled histology voxels. For our study we have developed and used a color based segmentation technique to identify and count positive staining without supervision. The resulting quantitative histology maps was than correlated with corresponding DBSI-derived maps.Materials and Methods
MRI of multiple sclerosis brain tissue: Three MS human brain tissues were examined using an Agilent DirectDrive console equipped with a 4.7 T magnet and a 15-cm inner diameter, actively shielded Magnex gradient coil. The tissues to be imaged was placed in a coil for data acquisition using the following parameters: repetition time 1s, spin echo time 43 ms, time between application of gradient pulse 25 ms, diffusion gradient on time 8 ms, slice thickness 0.5 mm, number of slices 1, field-of-view 6.4 x 6.4 cm2, number of average 1, data matrix 128 x 128. Diffusion sensitizing gradients were applied in 99 directions with max b-value = 3000 s/mm2.
Down-sampling high resolution histology images: Images were acquired with a Hamamatsu NanoZoomer 2.0-HT System (Hamamatsu) using a 40x objective and downsampled. At 40x magnification, the linear dimension of raw histology image pixels was 0.23 µm in contrast to 500 µm of MRI voxels. Thus, the raw histology image was down-sampled to match MRI voxel size, resulting in each down-sampled histology image voxel, containing 2174 x 2174 raw histology image pixels.
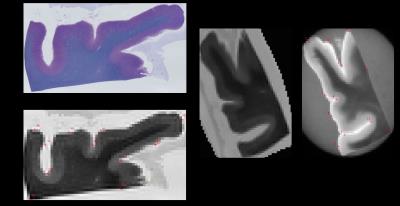
Histology Quantification: For this study, a color segmentation based method was used to detect and identify positive staining and counted objects of interest (axons /myelin) within the selected down-sampled histology voxels (Fig. 1A). For each down sampled voxel (Fig. 1B) regions of interest (ROI) (Fig. 1C) corresponding to the color of object of interest (myelin, blue for instance) were selected. From the selected ROIs, for each selected voxel(s), the resulting mask(s) (Fig. 1D) was generated by computing the “square root of sum of square distance in the voxel” with appropriately thresholding. The fraction of positive stain area in each voxel (Fig. 1E) is then computed, as the ratio between the number of positive staining pixels and the total number of pixels within the down sampled histology voxel and from this histology quantification map is generated (Fig 1F).
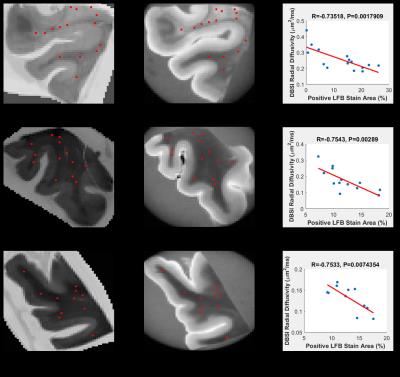
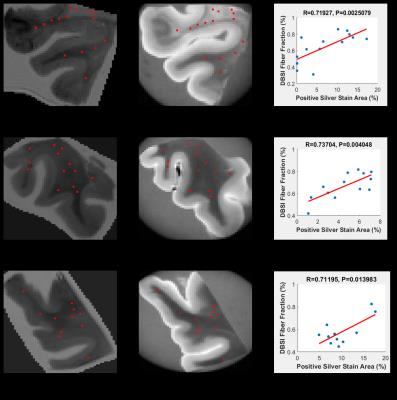
Co-registration between quantitative histology and DBSI maps: Non-linear registration was performed using the Medical Image Processing and Visualization tool. Landmarks along the perimeter of the tissue were manually placed on down-sampled histology (Fig. 2B) and diffusion weighted images (Fig. 2D), to compute the transformation function to match the DBSI maps with down-sampled histology maps. Through successful image co-registration, the selected voxels on the down-sampled histology maps can be transferred to MRI (Fig. 2C). For each autopsied human brain tissue, 11–15 down-sampled image voxels were randomly selected in brain white matter (red squares in Fig. 3A,B,C and 4A,B,C). These down-sampled image voxels represent regions of interest in the raw high-resolution histology images containing 2174 x 2174 native image pixels, i.e. equivalent of single MRI image voxel (red squares in Fig. 3D,E,F and 4D,E,F).
Results and Discussion
To show that DBSI metrics and quantitative histology findings measure the same biological characteristics, Spearman’s rank correlation coefficients were used to measure the strength of monotone increasing or decreasing association. For this study, we examined two of the primary pathologies, namely axonal loss and demyelination. The associations of interest were DBSI fiber fraction (reflecting residual axonal content) with positive silver stain, and DBSI radial diffusivity (reflecting myelin integrity) with positive LFB-PAS. For all three samples, DBSI-derived radial diffusivity negatively correlated with myelin content quantified by LFB-PAS staining (Fig. 3G,H,I), while DBSI-derived fiber fraction positively correlated with axonal content quantified by Bielschowsky’s silver staining (Fig. 4A,H,I). These findings are consistent with previous studies using DBSI1.Conclusion
In this study, we developed a method to quantify histological stained images based on color segmentation. The resulting quantitative histological maps generated were observed to be well correlated with corresponding DBSI maps.Acknowledgements
Supported in part by NIH R01-NS047592, P01-NS059560, and NMSS RG 5258-A-5, RG 1507-05315References
[1] Wang, Y., et al., Differentiation and quantification of inflammation, demyelination and axon injury or loss in multiple sclerosis. Brain, 2015. 138(Pt 5): p. 1223-38.Figures