2540
Optimization of Neurite Orientation Density and Dispersion Imaging (NODDI) for In Situ Imaging1Imaging Institute, The Cleveland Clinic, Cleveland, OH, United States, 2Mellen Center for Multiple Sclerosis, The Cleveland Clinic, Cleveland, OH, United States
Synopsis
In situ imaging is a valuable context for validation of imaging measures against histology. This contribution describes optimization of diffusion MRI for NODDI in in situ imaging of a multiple sclerosis patient. The results are expected to facilitate validation of advanced diffusion MRI and other tissue microstructure meausrements.
PURPOSE
NODDI has the potential to improve the specificity of diffusion MRI (dMRI) to cellular properties1. For example, the use of NODDI to measure the thickness of myelin sheaths2 provides a compelling rationale for validation against histology. Validation is typically performed on tissue from animal models or in fixed human tissue. However, the relevance of animal models to human disease and the impact of fixation on imaging can be unclear. In situ imaging avoids these problems by imaging recently-deceased human subjects prior to extraction and processing of tissue. To limit tissue degradation, the imaging must be completed within 6 hours of death. The purpose of this contribution is to optimize NODDI for in situ imaging, particularly with regard to overall scan time.METHODS
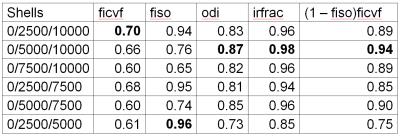
Informed consent was obtained from the family of one multiple sclerosis patient for inclusion in this internal review board-approved study. Imaging was performed within 4 hours of death on a Siemens Prisma with standard 20 channel head/neck array (Siemens Healthcare AG, Erlangen, Germany) with neck elements switched off. A multishell dMRI acquisition with total scan time of 31 minutes was performed (250mm x 250mm FOV, 100 x 100 matrix, 40 slices 2.5mm thick, TE = 105 msec, TR = 5800 msec, partial fourier factor 6/8, bandwidth 2380 Hz/Pixel. 32 b=0 scans, 64 volumes at b = 2500/5000/7500/10000 sec/mm2, 32 b=0 scans with reverse phase encoding). Postprocessing consisted of topup3,4 and eddy5 from FSL4. The NODDI toolbox (http://mig.cs.ucl.ac.uk/index.php?n=Tutorial.NODDImatlab) using MATLAB (MATLAB Release 2015a, The MathWorks, Inc., Natick, Massachusetts, United States) was used to calculate intracellular volume fraction (ficvf), isotropic volume fraction (fiso), orientation dispersion index (odi), isotropically restricted volume fraction (irfrac) and the product (1 – fiso)ficvf, which is used in the calculation of g-ratio2. Calculation of NODDI parameters was repeated using subsets of the full dataset consisting of b=0/2500/10000, 0/5000/10000, 0/7500/10000, 0/2500/7500, 0/5000/7500 and 0/2500/5000 sec/mm2. These subsets correspond to reduction in the overall scan time to 18 minutes.RESULTS
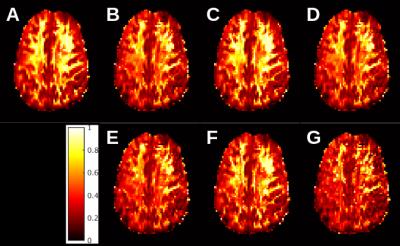
Figure 1 shows maps of the product (1 – fiso)ficvf for the full acquisition and the shortened acquisitions. Inspection suggests that the acquisition using b=0/5000/10000 sec/mm2 most closely matches the full acquisition. Table 1 summarizes pearson correlation coefficients for voxel-by-voxel correlations between the full acquisition and each of the subsets for each of the NODDI parameters.DISCUSSION
Overall, the sub-acquisition with b=0/5000/10000 sec/mm2 has the highest fidelity to the full acquisition. This can be seen from both qualitative inspection, exemplified by figure 1, and from inspection of the correlation coefficients. Although correlations are, overall, high, the acquisition using b=0/5000/10000 sec/mm2 has the highest overall correlation for odi, irfrac and (1 – fiso)ficvf. As the value of (1 – fiso)ficvf is a key part of the calculation of g-ratio, high correlation for this quantity is particularly important. Of note is the need for large b-values. For in vivo measurements, the maximum b-value is typically 3000 sec/mm2 1. The need for large b-values is due to reduced diffusivity even prior to fixation. This finding agrees diffusion tensor imaging performed in situ6.CONCLUSION
These results are an initial step towards validation of NODDI against histology using in situ imaging. Limiting the scan time of the dMRI acquisition permits acquisition of images using other complementary modalities that quantify myelin and T1. Although diffusivity is reduced as compared to live tissue, it is unclear if this will pose a problem for comparison against histological measures of axonal density. Future work will focus on such validation.Acknowledgements
We gratefully acknowledge support from NIH R01NS091683.References
1. Zhang, H., Schneider, T., Wheeler-Kingshott, C. A. & Alexander, D. C. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 2012; 61(4):1000-1016.
2. Stikov, N. et al. In vivo histology of the myelin g-ratio with magnetic resonance imaging. Neuroimage 2015; 118(397-405.
3. Andersson, J. L., Skare, S. & Ashburner, J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage 2003; 20(2):870-888.
4. Smith, S. M. et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 2004; 23 Suppl 1(S208-219.
5. Andersson, J. L. & Sotiropoulos, S. N. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage 2016; 125(1063-1078.
6. Scheurer, E. et al. Forensic application of postmortem diffusion-weighted and diffusion tensor MR imaging of the human brain in situ. AJNR Am J Neuroradiol 2011; 32(8):1518-1524.
Figures