2440
Patient-specific 3D Printable Anatomical Brain Models from a Web App1Translational Imaging Group, CMIC, Dep. of Medical Physics and Biomedical Engineering, University College London, London, United Kingdom
Synopsis
The process for obtaining 3D printed models from patient-specific brain Magnetic Resonance Imaging (MRI) datasets remains tedious for non-computer scientists, despite the availability of various open-source image segmentation software and the current affordability of 3D printers. In this work, we present a web app that takes advantage of cloud computing technologies to propose a practical and straigntforward system for anyone to upload brain MRI datasets and automatically receive corresponding patient-specific 3D printable models. Using our tool requires absolutely no specific installation or configuration.
Introduction
Traditionally, anatomical models have been used as training or teaching tools in clinical environments and for medical imaging research. More recently, advances in segmentation algorithms and increased availability of 3D printers have eased the creation of cost-efficient patient-specific models. However, the process for obtaining 3D printed models from patient-specific Magnetic Resonance Imaging (MRI) datasets remains tedious for non-computer scientists as it typically requires the use of many different software tools and a substantial amount of manual intervention. The typical pipeline for obtaining such patient-specific physical models is composed of three main steps: image segmentation, mesh refinement and 3D printing.
We leverage the use of cloud computing technologies to propose a practical and straightforward system for anyone to upload brain MRI datasets and automatically receive corresponding patient-specific 3D printable models. Using our tool requires absolutely no specific installation or configuration.
Methods
We present a new pipeline to compute, on the cloud, patient-specific stereolithography files (STL) ready for 3D printing from MRI scans of the brain provided in the NIFTI file format. Users can upload an MRI scan of the brain to our web platform, NiftyWeb1, and, using Geodesic Information Flows (GIF)2, our system automatically segments the specified regions of interest, converts the segmentation to a mesh model and refines it for 3D printability (see Figure 1). Finally, a link to download the resulting STL file is sent to the user via email.
We have integrated our pipeline within NiftyWeb (http://cmictig.cs.ucl.ac.uk/niftyweb), our web portal for publicly disseminating and making available image analysis tools with minimal effort and with optimal algorithm configuration. NiftyWeb provides a user-friendly interface to upload image datasets without the need to create a dedicated account.
For image segmentation, we rely on GIF2 which is a segmentation technique that uses a template database as sources of information and is able to propagate voxel-wise annotations, such as tissue segmentation or parcellation, between morphologically dissimilar images by diffusing and mapping the available examples through intermediate steps, even in the presence of large-scale morphological variability. The template database used for this study has 95 MRI brain scans neuroanatomically labelled according to the Neuromorphometrics protocol. GIF is available either as standalone open-source software inside NiftySeg (http://niftyseg.sf.net) or as a dedicated Software-as-a-Service (SaaS) tool as part of NiftyWeb. Note that the current SaaS version of GIF does not directly provide 3D printable models.
In the proposed pipeline, the segmentation obtained from GIF is automatically refined into a 3D printable mesh by using two publicly available filters from the Visualization Toolkit (VTK)3. First, the iso-surface of the model is detected using vtkContourFilter. Second, in order to remove spurious contours, the iso-surface model is refined using vtkWindowedSincPolyDataFilter4, which evenly distributes the location of the mesh vertices using a windowed cardinal function interpolation kernel.
For the segmentation of the hippocampus and whole brain, one healthy volunteer was scanned using a 3T Philips scanner. A 3DT1-weighted acquisition with a voxel resolution of 1×1×1mm3 was acquired and a written informed consent was obtained from the participant.
We used an Ultimaker 2 (Ultimaker, Chorley, England) FDM printer to create our 3D physical models. They were prepared for the printer using the slicing software Cura, which is provided for free by Ultimaker. All models were printed with a layer height of 0.12 mm and a shell, bottom, and top thickness of 0.8 with a nozzle size of 0.4 mm. The material used for printing was "enhanced Polymax" polylactic acid (PLA) (PolyMax; Polymakr, Changshu, China).
Results
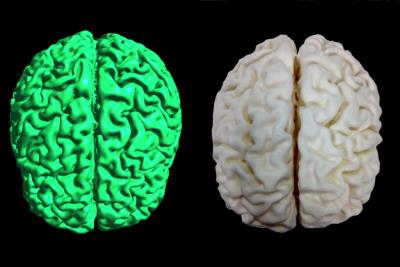
We computed and 3D printed two 3D physical models using the proposed pipeline to create patient-specific anatomical models: one is a whole brain model and the other a hippocampus model (see Figure 2 and 3). The 3D prints were created at 20% infill. The printing durations for the hippocampus and whole brain were 1.5h, and 67.5h respectively. Figure 2 and 3 show direct comparisons between the mesh and the 3D printed models. It illustrates the good quality and precision of the 3D printed models.Conclusions
In this work, we have introduced a new web app, as part of our web portal for medical image cloud computing NiftyWeb, for automatically creating 3D printable models on the cloud. The tool computes patient-specific 3D printable models for any combination of the 144 available brain areas thereby avoiding the need of manual editing, post-processing, mesh refinement or software installation. The platform eases access for any user to obtain patient-specific 3D printable models for teaching, surgical planning or building research phantoms amongst other applications.Acknowledgements
NIHR BRC UCLH/UCL High Impact Initiative (BW.mn.BRC10269), EPSRC (EP/H046410/1; EP/J020990/1; EP/K005278; EP/K503745/1; NS/A000027/1), MRC (MR/J01107X/1), Wellcome Trust (WT101957; 106882/Z/15/Z) and Brain Research Trust.References
1) F. Prados, M. J. Cardoso, N. Burgos, C.AM Wheeler-Kingshott, S. Ourselin (2016). NiftyWeb: web based platform for image processing on the cloud. International Society for Magnetic Resonance in Medicine (ISMRM) 24th Scientific Meeting and Exhibition - Singapore 2016
2) M. J. Cardoso, M. Modat, R. Wolz, A. Melbourne, D. Cash, D. Rueckert, and S. Ourselin (2015), Geodesic Information Flows: Spatially-Variant Graphs and Their Application to Segmentation and Fusion. IEEE TMI vol. 34(9), 1976-88
3) W. Schroeder, K. Martin, B. Lorensen, (2006), The Visualization Toolkit (4th ed.), Kitware, ISBN 978-1-930934-19-1
4) G. Taubin and T. Zhang (1996) Optimal surface smoothing as filter design. Computer Vision - ECCV '96: 4th European Conference on Computer Vision Cambridge, UK, April 15-18, Springer Berlin Heidelberg, 283-292 ISBN 978-3-540-49949