2439
A patch-based method for lesion in-painting in the spinal cord1Translational Imaging Group, CMIC, Dep. of Medical Physics and Biomedical Engineering, University College London, London, United Kingdom, 2UCL Institute of Neurology, Queen Square MS Centre, University College London, London, United Kingdom, 3Department of Brain and Behavioural Sciences, University of Pavia, Italy, 4Brain MRI 3T Mondino Research Center, C. Mondino National Neurological Institute, Italy
Synopsis
Multiple Sclerosis lesions can impair most automated image-processing pipelines. They can be present with different sizes, contrasts and shapes and at multiple locations across the central nervous system. Whilst lesion-filling is often a required pre-processing step in the analysis of brain MR images, its potential utility in spinal cord imaging remains unexplored. This study introduces a method for in-painting lesions in the spinal cord and demonstrates its efficacy for improving the results of spinal cord MR image analysis.
Introduction
Multiple Sclerosis (MS) lesions can be present anywhere along the central nervous system and can have a major impact on image processing pipelines utilized in brain and spinal cord (SC) imaging. Lesions are of different shapes, sizes and contrast in different subjects and can morph over time. Their presence introduces bias/errors in common MR image analysis pipelines such as segmentation, non-rigid registration, voxel-based morphometry and atrophy measurements. In brain imaging, the impact of lesions on image processing pipelines is reduced by means of in-painting during pre-processing1,2. However, a similar method is not yet established for SC imaging.
This study validates and translates a cutting-edge lesion filling method for in-painting MS lesions in the context of SC imaging2.
Methods
We adapted a multi-time-point, modality-agnostic, patch-based method for lesion-filling in the brain2 to SC imaging. MR images of the SC typically have high in-plane resolutions, but large slice thicknesses and, for this reason it is common to apply processing in a slice-by-slice manner. However, trying to fill lesions in a slice-by-slice manner with a patch-based method is challenging due to the presence of large lesions and the small cross-sectional size of the SC, resulting in the scarcity of potentially matched healthy patches. In order to overcome this limitation, we have adapted the method to work with 2D patches, by searching for similar 2D patches in 3D volumes of interest, thus increasing the number of potential good patches for in-painting a given voxel. As the method works with adaptive patch sizes2, the 3D volume of interest can include a variable number of slices depending on the size and the location of the SC lesion that needs to be filled.
We analysed data from 25 healthy subjects and 33 MS patients with visible lesions in the SC (lesion load mean(std) = 146.19 (66.33) ml). MRI data were obtained using a 3T Philips Achieva scanner using a 3D-FFE sequence (0.5x0.5x5 mm3), with the center of the volume positioned at the C2-3 intervertebral disc. Lesions were manually drawn by a rater using JIM v6 (Xinapse Systems, UK).
In order to assess the inpainting performance of the lesion-filling method, we segmented the cross-sectional area (CSA) and the grey matter (GM)3 using non-filled (RAW), and filled images and compared the masks obtained from the segmentation pipelines with the consensus masks of three raters who manually outlined GM and semi-automatically outlined CSA on the non-filled images, also using JIM.
In order to segment the CSA and the GM, we used a new, fully automated SC segmentation technique3 that incorporates two different multi-atlas segmentation propagation and fusion techniques: OPAL4 and STEPS5. The template library needed for the label fusion was comprised of labeled images from 25 healthy subjects. For each subject there were three slices, with the middle slice centered at C2-3 level. In order to maximise the size of the library, all the scans were left-right flipped, resulting in a final template library of 150 2D slices (25 datasets * 3 slices * 2 L/R flip). For each image in the template library, the corresponding consensus CSA and GM were provided by the three experienced raters.
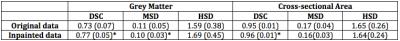
Quantitatively, we computed the Dice score coefficient (DSC), the Hausdorff surface distance (HSD) and the Mean surface distance (MSD) between the automatic and the consensus cord segmentation. Statistical differences in parameters were assessed using paired t-tests.
Results
We obtained improved segmentation results using filled SC images when compared to un-filled images (Figure 1 and Table 1). The spatial agreement between masks (DSC) significantly improved for the CSA and the GM (p<0.05) when segmentations were performed using in-painted images. The agreement between the mask boundaries were also significantly improved (MSD, p<0.05) for the GM after lesion filling. On the other hand, WM contours were less affected by the presence of lesions due to its oval shape and the choice of segmentation technique. Global error changes (HSD) were not significant for either the GM or WM.Conclusions
This study introduced a technique for filling MS
lesions in SC images and found significant improvement in GM and CSA segmentations
of the SC with MS lesions when lesions were filled. Future work will explore
the performance of the method using multi-modality images, images with more slices,
as well as other locations along the SC.Acknowledgements
NIHR BRC UCLH/UCL High Impact Initiative (BW.mn.BRC10269), EPSRC (EP/H046410/1,EP/J020990/1,EP/K005278), MRC (MR/J01107X/1), UK MS Society, ISRT, WFL, CHNF (INSPIRED) and Brain Research Trust.References
1) D.T. Chard, J.S. Jackson, D.H. Miller, C. Wheeler-Kingshott. Reducing the impact of white matter lesions on automated measures of brain gray and white matter volumes. J. Magn. Reson. Imaging, 32 (2010), pp. 223–228
2) F. Prados, M.J. Cardoso, B. Kanber, O. Ciccarelli, R. Kapoor, C. Wheeler-Kingshott, S. Ourselin. A multi-time-point modality-agnostic patch-based method for lesion filling in multiple sclerosis. NeuroImage, 139 (2016), pp. 376-384
3) F. Prados, M.J. Cardoso, M.C. Yiannakas, L.R. Hoy, E. Tebaldi, H. Kearney, M.D. Liechti, D. H. Miller, O. Ciccarelli, C. Wheeler-Kingshott, S. Ourselin. Fully automated grey and white matter spinal cord segmentation. Scientific Reports, (2016).
4) R. Giraud, V. Ta, N. Papadakisc, J. V. Manjón, D. L. Collins, P. Coupé, Alzheimer's Disease Neuroimaging Initiative. An Optimized PatchMatch for multi-scale and multi-feature label fusion. NeuroImage, 124A (2016), pp. 770-782
5) M.J. Cardoso, K.K. Leung, M. Modat, S. Keihaninejad, D. Cash, J. Barnes, N.C. Fox, S. Ourselin. STEPS similarity and truth estimation for propagated segmentations and its application to hippocampal segmentation and brain parcelation. Med. Image Anal., 17 (2013), pp. 671–684
Figures